Keywords
GI pathogens, respiratory pathogens, nutritional status, gut-lung axis
GI pathogens, respiratory pathogens, nutritional status, gut-lung axis
Lower respiratory infections (LRI) and diarrhoeal diseases are the top leading preventable causes of mortality and morbidity globally in children under five years of age, and are the annual cause of 12% (700,000) and 9% (446,000) of deaths, respectively1–3. Children with frequent and recurrent infections are at risk of malnutrition which also predisposes them to further infection4. Malnutrition is one of the most important risk factors for both diarrhoeal disease and LRI3,5, and is associated with about half of all under-five deaths6. Reducing the burden of malnutrition could, therefore, concomitantly decrease respiratory and diarrhoeal disease amongst high-risk children. This was estimated in the “Global Burden of Disease” study to be a reduction of 9% of LRI and 12% of diarrhoeal disease over the past three decades1,3.
Recently, large-scale studies including the “Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and Consequences for Child Health and Development” (MAL-ED) study and the “Global Enteric Multi-site” (GEMS) study explored the association between gastrointestinal (GI) pathogens, malnutrition and gut function with other long-term effects to understand the pathophysiology of malnutrition7–9. These studies noted that subclinical infections and quantity of pathogens are negatively associated with linear growth, and that this persists until two years of age8–10. Cumulative insults of infections like diarrhoeal on children likely cause failure of catch-up growth, resulting in growth faltering and decreased cognitive development11–14. However, the association between these pathogens and respiratory infections and short-term growth has not been explored.
The gut and lungs both have the same embryonic origin. Thus, while the mechanisms are not understood, studies have shown that the two sites interact in health and disease. Specifically, animal studies have demonstrated how the ‘gut-lung axis’ of host-associated gut and respiratory microbiota appear to influence local and systemic immunity15–18. However, while these studies have focused on how gut microbiota is broadly protective against respiratory infection19, there has been less attention paid to the reverse relationship, i.e. respiratory microbiota and pathogens influencing the gut, and subsequent association with growth. Studies have indicated a link between respiratory infections and growth20,21 although generally the pathways between growth, nutrition and infection are likely bidirectional.
Cryptosporidium infection is a common cause of diarrhoeal infection, malnutrition and excess mortality amongst children in developing countries9,10. The objective of this study was to describe detection of diarrhoeal and respiratory pathogens in children who were hospitalised with diarrhoeal disease (with detection of Cryptosporidium) at a tertiary hospital in Malawi, and to examine the association with short-term growth over an eight-week follow-up period.
This is a secondary data analysis from a prospective longitudinal observational study evaluating respiratory cryptosporidiosis in paediatric diarrhoeal disease22,23. The main study recruited children presenting with primary gastrointestinal (GI) symptoms to Queen Elizabeth Central Hospital in Blantyre, Malawi, from March 2019 to April 2020. Children were eligible to participate if they were two-24 months of age and had at least three or more loose stools within the past 48 hours and lived 15km outside of the study22,23. Children were excluded if they had visible blood in loose stools, or dysentery24. Parents provided written informed consent. The study was approved by the University of Malawi College of Medicine Research Ethics Committee (P.07/18/2438) and the Liverpool School of Tropical Medicine Research Ethics Committee (18-066).
Participants positive for Cryptosporidium spp. in either respiratory or GI tract specimens at enrolment were followed up every two weeks until eight weeks post-enrolment. At each visit, history and physical exam were conducted, and induced sputum and stool were collected. The induced sputum procedure has been previously described in detail elsewhere22,23. In brief, sputum samples were obtained via oropharyngeal suctioning after nebulized 3% sodium chloride treatment and processed for microscopy and multiplex PCR testing.
For the main study, children with a positive PCR for Cryptosporidium in any of the collected samples were followed up, and they constitute the sample included in this sub-analysis23. At enrolment, Cryptosporidium spp. in stool/sputum/ NP were detected using PCR analysis specifically for Cryptosporidium spp. Diarrhoea was defined as ≥3 loose stools within the past 48 hours. A diarrhoeal episode was termed symptomatic if the participant reported any GI symptoms (to include abdominal pain/tenderness, dehydration, vomiting, and/or poor feeding) and the stool sample collected was PCR-positive for any pathogen, and asymptomatic if the participant did not report any GI symptoms but the stool was PCR-positive for any pathogen. Respiratory symptoms included cough, runny nose, difficulty in breathing, wheezing, chest indrawing/retractions, and/or crackles.
Nutrition indices were defined according to WHO growth standards25. We defined wasted, underweight or stunted, as weight-for-length z score (WLZ), weight-for-age z scores (WAZ) and length-for-age z score (LAZ) <-2, respectively.
We extracted DNA from stool samples using a QIAamp Fast DNA Mini Kit (Qiagen, Hilden, Germany) with a procedure modified from that of the manufacturer as previously described26. Briefly, 200mg solid stool or 200µL liquid fecal samples were first mixed with InhibitEX buffer and glass beads before bead beating (Tissue Lyser II, Qiagen). Resulting lysates were heated at 95°C for 5 minutes prior to proceeding according to the manufacturer’s protocol. Sputum samples for Cryptosporidium detection were extracted using QIAamp DNA mini kit. Briefly the 180µl ATL buffer and 20ul proteinase K was added to 300ul sample and incubated at 56°C for 1–3 hours with occasional vortexing during the incubation. This was followed by addition of 200µl Buffer AL and the sample was incubated at 70°C for 10 minutes. 200ul of absolute ethanol was added and this was followed by washing using 500µl buffer AW1 and 500µl buffer AW2. DNA was eluted using 200µl of elution buffer.
All samples were spiked with Phocine herpes virus (PhHV) and MS2 phage to be used as extraction controls. One extraction blank (200µL nuclease-free water as the sample) was included in each batch of extractions to monitor for contamination.
We performed qPCR as previously described27. These qPCRs were carried out using the ViiA7 or QuantStudio 7 Flex Real-Time PCR instruments (Thermo Fisher, Waltham, MA, USA). Primers (Crypto F primer: GGGTTGTATTTATTAGATAAAGAACCA, Crypto R primer: AGGCCAATACCCTACCGTCT) and probe (<FAM>GTGACATATCATTCAAGTTTCTGAC<BHQ1>). These were sourced from Integrated DNA Technologies (IDT, Coralville, Iowa, USA) and Sigma (Sigma-Aldrich, Haverhill, UK). All resulting qPCR data were analyzed using QuantStudio 6 and 7 Flex Real-Time PCR System Software, version 1.3 (Thermo Fisher). For initial denaturation and Taq activation we used one cycle at 95°C for 3 minutes, for amplification and subsequent target detection we used a total of 40 cycles (95°C for 10 seconds and 60°C for 1 minute). An analytical cutoff of 35 cycles was applied to the data (i.e. Ct values ≥35.0 were considered negative).
In sputum, multiplex testing detected bacteria (S. pneumoniae, Staphylococcus aureus, M. catarrhalis, Bordetella pertussis, Haemophilus influenzae and H. influenzae type b, Chlamydia pneumoniae, Mycoplasma pneumoniae, Klebsiella pneumoniae, Legionella pneumophila, and Salmonella species); viruses (influenza A/B/C, RSV A/B, parainfluenza virus types 1-4, coronaviruses NL63, 229E, OC43, and HKU1, human metapneumovirus A/B, rhinovirus, adenovirus, enterovirus, parechovirus, bocavirus, cytomegalovirus); and parasites (Pneumocystis jirovecii). A cycle threshold (Ct) of <38 was considered as a positive for any of these pathogens. Cryptosporidium spp. detection of respiratory specimens were measured using quantitative polymerase chain reaction (qPCR), with a positive result corresponding to a Ct <35.
In stool, GI pathogens were detected using qPCR in a TaqMan Array Card (Thermo Fisher, Waltham, MA) using a custom design developed at the Houpt Laboratory (Charlottesville, VA [25]). These were done at week 2 to week 8. Multiplex testing detected rotavirus, norovirus GII, adenovirus, astrovirus, sapovirus, enterotoxigenic Escherichia coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), Shiga-toxigenic E. coli (STEC), Shigella/enteroinvasive E. coli (EIEC), Salmonella, Campylobacter jejuni/coli, Vibrio cholerae, Clostridium difficile, Cryptosporidium spp, C. parvum, C. hominis, Giardia lamblia, Entamoeba histolytica, Ascaris lumbricoides, and Trichuris trichiura). We considered a pathogen as present if Ct was <35 in all pathogens. For pathogens that were positive and are associated with diarrhoea in children under five years old8,13, we calculated the prevalence of diarrhoeagenic Ct cut-offs (diarrhoea-associated Ct quantity) based on the GEMS study to estimate the prevalence of diarrhoeal samples in this population27. An increased pathogen load was defined as at least three pathogens present in each sample per participant per study visit to compare how these relate with other demographics28.
At enrolment, categorical variables were compared using Pearson’s X2 test or Fisher’s exact test. Continuous variables were compared using Student’s t-tests or nonparametric Mann-Whitney U tests where data were nonnormally distributed. We presented diarrhoeagenic quantitative cut-off based on the GEMS study to estimate the burden of diarrhoea attributed to the common causes of diarrhoeal pathogens in this population. These cut-off values are useful in studies that have no controls or that do not have diarrhoea data like our study. Comparison for different characteristics across the eight-week study period was done using one-way repeated measures analysis of variance and mixed-effect model analysis. Statistical significance was set at 0.05, characteristics that showed a significant change over the follow-up period were included in a mixed-effect model analysis as confounders. Statistical analysis was performed using Stata software, version 16 (StataCorp. 2019. College Station, TX, USA).
From March 2019 to April 2020, 755 children were screened and 162 were recruited into the study. Of the 162 enrolled, 37 (23%) were positive for Cryptosporidium spp., 36 were entered into follow-up, and 27 children (75%) completed the 8-week follow-up, which was discontinued early due to COVID-19. Of these, the median age was 5.5 (IQR 2,14) months and 18 (64%) were male. Only 1 (3%) was HIV-infected, but HIV status was unknown in over half of the children (20/27). The enrolled study population is described elsewhere29.
We tested 104 stool and sputum samples from the 27 participants that had completed follow-up from week 2 to week 8 post-enrolment. At least one pathogen was detected in all the 104 stool samples, while 87/104 (84%) of the sputum samples had at least one pathogen detected. Amongst the 104 stool samples collected, diarrhoeagenic E. coli was the most abundant pathogen (92/104 [89%]), of which EAEC was the most common subtype (88/92 [95%]) followed by typical EPEC (42/92 [45%]). Cryptosporidium spp. (60/104 [(57.6%]) were the second most common stool pathogen, of which a third were C. hominis (20/55), one sample was C. parvum and the rest were not identified. Adenovirus pan (44/104 [42%]),
Campylobacter pan (43/104 [41%]), Campylobacter jejuni/coli (34/104 [33%]), rotavirus (36/104 [34%] and norovirus (34/104 [32%]) were the other common pathogens identified. Over half (58/104 [56%]) of the follow-up stool samples with a pathogen associated with diarrhoea were below the diarrhoeagenic cut-offs as presented in the GEMS study (Figure 1). Out of the 104 induced sputum samples, S. pneumoniae was the most abundant pathogen 87/104 (84%) followed by Human Rhinovirus (58/104 (56%)), M. catarrhalis (52/104 [50%]), Adenovirus (29/104 [28%]) and H. influenzae (21/104 [20%]). The majority of stool samples (80/104 [77%]) and almost half of sputum samples (49/104 [47%]) had at least three pathogens detected (data not shown). Of the 49 sputum samples that had a high pathogen load, 42/49 (85%) simultaneously had ≥3 stools pathogens, although this was not statistically significant.
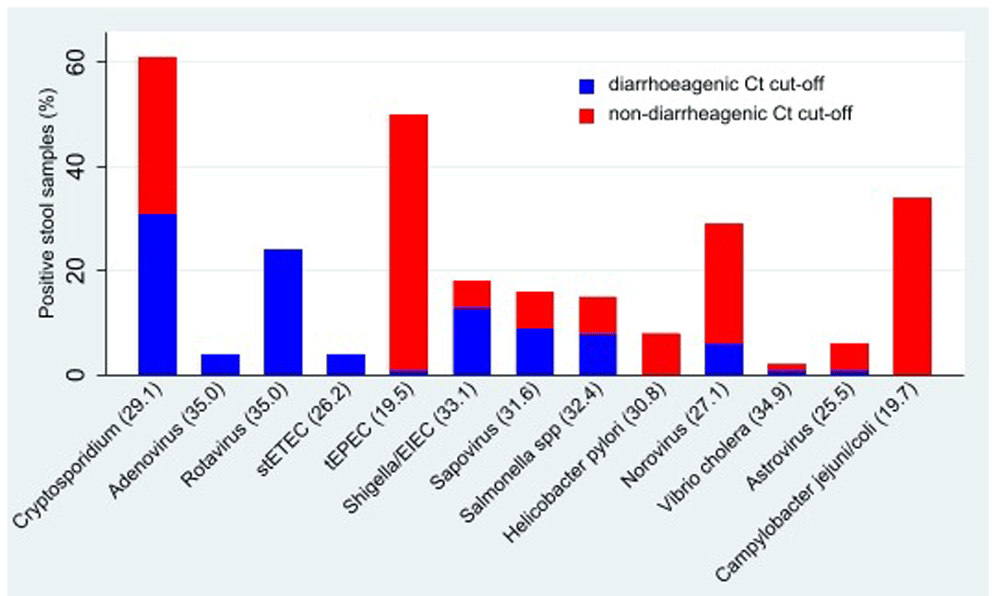
stETEC - heat-stable enterotoxin-producing E. coli; tEPEC – typical enteropathogenic E. coli; EIEC – enteroinvasive E. coli; H. pylori –Helicobacter pylori; V. cholera- Vibrio cholera.
GI pathogens were detected in all stool samples from two weeks to eight weeks post-enrolment. In contrast, respiratory pathogens were detected in all sputum samples at 2 weeks and in 20/25 (80%) samples at the end of the eight weeks. On average, there were 5.1 (SD 2.1) stool pathogens detected per participant per study visit over the follow-up period, while an average of 3.5 (SD 1.8) sputum pathogens were detected per participant per study visit over the follow-up period (Table 1). There was an average of 3.1 (SD 1.6) bacteria compared to 1.0 (SD 0.8) parasites and 1.0 (SD 0.9) viruses in stool samples collected, and an average of 2.0 (SD 1.1) bacteria, 1.4 (SD 1.1) viruses and 0.3 (SD 0.5) parasites in sputum samples (Figure 2).
| Characteristics | Study period | p-value | |||||
|---|---|---|---|---|---|---|---|
| Enrolment* | 2w | 4w | 6w | 8w | Total | ||
| Weight-for-age z score, mean (SD) | -1.2 (0.9) | -0.9 (1.0) | -0.8 (1.0) | -0.8 (0.9) | -0.8 (0.9) | - | 0.002 |
| Length-for-age z score, mean (SD) | -1.8 (1.4) | -1.7 (1.3) | -1.4 (1.4) | -1.5 (1.3) | -1.4 (1.1) | - | 0.204 |
| Weight-for-length z score, mean (SD) | -1.2 (0.9) | -1.2 (1.4) | -0.2 (1.2) | -0.0 (1.3) | -0.1 (1.2) | - | 0.673 |
| Total number of respiratory pathogens detected/participant/visit (mean, SD)) | - | 3.7 (1.6) | 3.6 (1.8) | 3.6 (1.8) | 2.8 (1.8) | 3.5(1.8) | 0.115 |
| Mean number of bacterial respiratory pathogens/visit (N=202) | - | 2.2 (1.0) | 1.8 (1.1) | 1.7 (1.0) | 2.1 (1.2) | 2.0(1.1) | 0.347 |
| Mean number of viral pathogens per visit (N=141) | - | 1.4 (1.1) | 1.6 (1.0) | 1 (1.0) | 1.5 (1.1) | 1.4(1.1) | 0.148 |
| Mean number of parasitic pathogens per participant/visit (N= 48) | - | 0.5 (0.6) | 0.5 (0.7) | 0.3 (0.5) | 0.3 (0.4) | 0.3(0.5) | 0.343 |
| Visit to health centre with respiratory symptoms in the past 7 days (%) | - | 10/27 (37%) | 6/24 (25%) | 5/27 (18%) | 7/24 (29%) | - | 0.107 |
| Cough | - | 5/10 (50%) | 2/6 (33%) | 2/5 (40%) | 5/7 (71%) | - | 0.147 |
| Rhinorrhoea | - | 7/10 (70%) | 4/6 (66%) | 1/4 (20%) | 3/7 (43%) | - | 0.347 |
| Visit to a health facility for a diarrhoea episode in past 7 days | 0/24 (0%) | 2/ 24 (8%) | 4/27 (15%) | 3/24 (13%) | 0.141 | ||
| Total number of diarrhoea pathogens detected/participant/visit (mean, SD) | - | 5.3 (2.0) | 5.1 (2.1) | 4.7 (2.1) | 5.2 (2.5) | 5.1 (2.1) | 0.817 |
| Number of bacterial GI pathogens detected/ participant/visit (mean, SD)d (N=320) | - | 2.8 (1.6) | 3.1 (1.8) | 3.1 (1.4) | 3.4 (1.8) | 3.1 (1.6) | 0.375 |
| Number of viral GI pathogens detected/ participant/visit (mean, SD)e (N=10) | - | 1.3 (1.0) | 1.0 (1.0) | 0.8 (1.0) | 0.8 (1.0) | 1.0 (0.8) | 0.087 |
| Number of parasitic GI pathogens detected/ participant/visit (mean, SD)f (N=98) | - | 1.2 (0.7) | 1.0 (0.9) | 0.7 (0.8) | 1.0 (0.9) | 1.0 (0.8) | 0.099 |
GEMS, Global Enteric Multicenter Study; GI, gastrointestinal; SD, standard deviation; w, week
*TaqMan Array was only run at follow up visits
aS. pneumoniae, S. aureus, M. catarrhalis, B. pertussis, H. influenzae and H. influenzae type b, C. pneumoniae, M. pneumoniae, K. pneumoniae, L. pneumophila, and Salmonella species
bInfluenza A/B/C, RSV A/B, parainfluenza virus types 1–4, coronaviruses NL63, 229E, OC43, and HKU1, human metapneumovirus A/B, rhinovirus, adenovirus, enterovirus, parechovirus, bocavirus, cytomegalovirus
cCryptosporidium, P. jirovecii
dEnterotoxigenic Escherichia coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), Shiga-toxigenic E. coli (STEC), Shigella/enteroinvasive E. coli (EIEC), Salmonella, Campylobacter jejuni/C. coli, Vibrio cholerae, Clostridium difficile
eRotavirus, norovirus GII, adenovirus, astrovirus, sapovirus
fCryptosporidium, Giardia lamblia, Entamoeba histolytica, Ascaris lumbricoides, and Trichuris trichiura
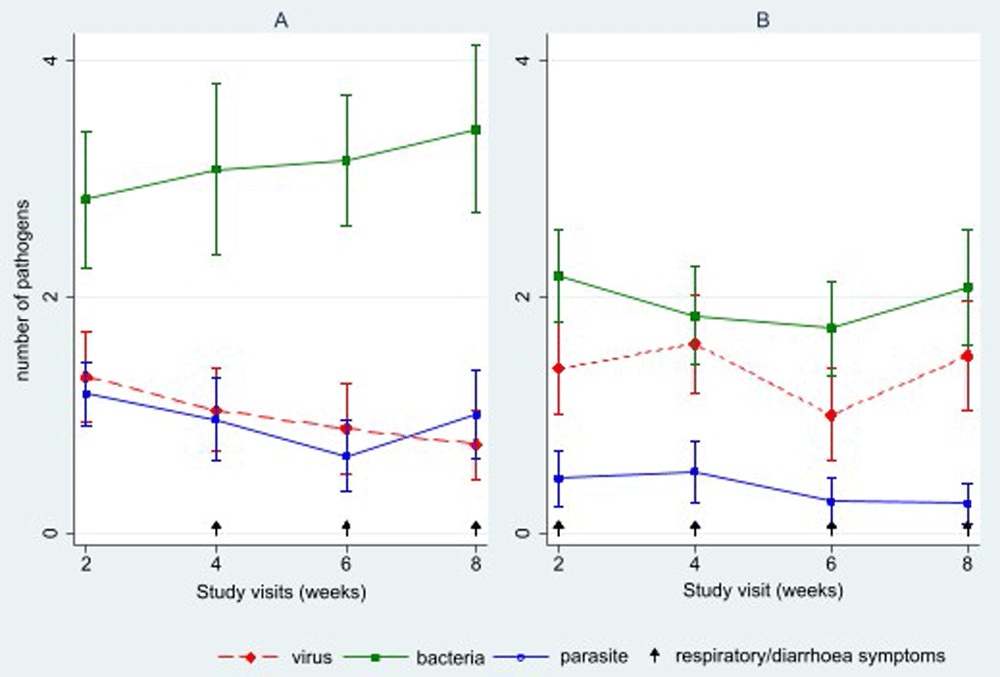
A) Stool B) Sputum.
Figure 3A shows the changes in WAZ, WLZ and LAZ across the study period. Participants had low LAZ and WAZ at enrolment, and the average change in WAZ, LAZ, and WLZ scores over the eight-week period were 0.5 (0.6), 0.4 (1.4) and 0.4 (1.4), respectively. There was a significant change in WAZ across the follow-up period (p=0.002), but no significant changes were seen in LAZ and WLZ. Children with ≥3 GI pathogens in a sample had lower LAZ compared to those with <3 pathogens at two weeks (-2.0±1.1 vs 0.1±0.5), four weeks (-1.6±1.4 vs -0.5±0.8), six weeks (-1.8±1.4 vs -1.0±1.2) and eight weeks (-1.6±1.2 vs -0.6±0.9), and this was statistically significant (Figure 3B). This was not noted for WLZ and WAZ scores. No obvious changes in WLZ, WAZ and LAZ were noted with respiratory pathogen detection over the study period (Figure 3C). There was also no difference in any of the nutritional indices amongst children with ≥3 of both respiratory and GI pathogens compared to those with <3 (Figure 3D).
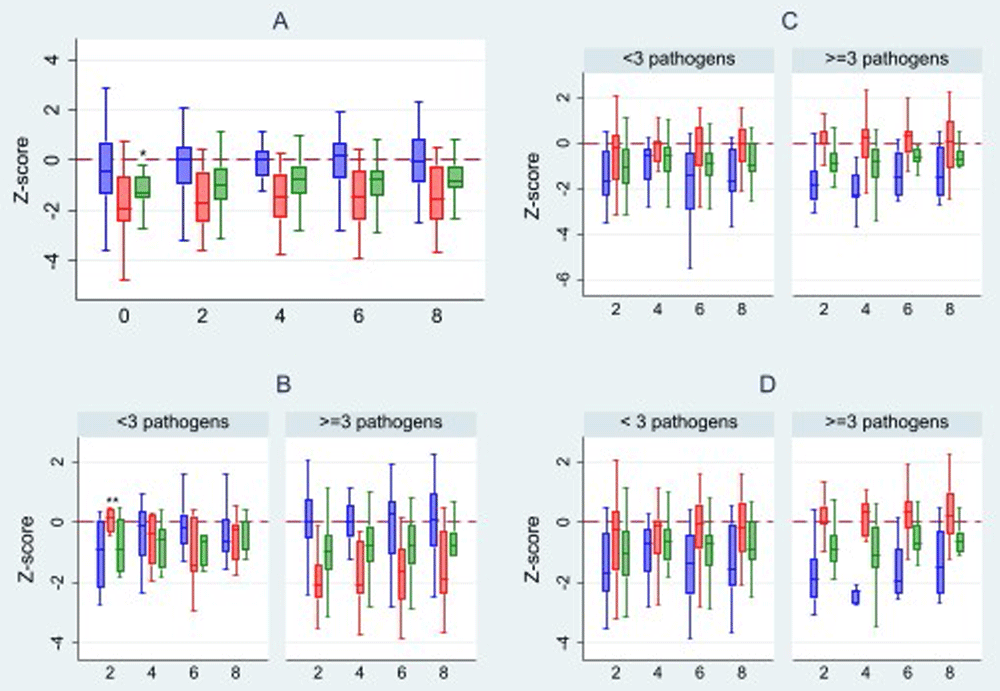
A) overall; B) in relation to average GI pathogens of i) <3 and ii) ≥3; C) in relation to average respiratory pathogens of i) <3 and ii) ≥3. D) in relation to respiratory and GI pathogens *p-value 0.002 **p-value 0.001  WLZ score
WLZ score  LAZ score
LAZ score  WAZ score
WAZ score
This is the first description, to our knowledge, of both respiratory and GI pathogens in young children in a low- and middle-income country and association with short-term growth in the eight weeks after hospitalisation with diarrhoea. We found that our population had low anthropometric indices, and that these indices showed minimal change over the eight weeks after hospitalisation. A high average number of GI pathogens was detected throughout the eight weeks, and this was associated with GI symptoms. A high average of respiratory pathogens was also detected throughout the eight weeks, predominantly without associated respiratory symptoms. Significant changes were only noted in WAZ and not the other anthropometric measures over the eight-week follow-up period. Additionally, participants with ≥3 GI pathogens had a lower mean LAZ score at all follow-up visits. Having a high number of both respiratory and GI pathogens was not associated with changes in nutritional indices over the follow-up period.
Previous studies that have evaluated GI pathogens in young children after hospitalisation and association with short-term growth have typically focused on a single pathogen12,30–32. However, co-infection with GI pathogens is common amongst children under two years and has significant effects on growth compared to single pathogens33–36. Short term growth after an infection in children under two years old is important because it allows for catch-up growth in this critical period for children to attain their optimal weight and height increment over time11,37. However, catch-up growth typically occurs in the absence of diarrhoea, whether clinical or subclinical14,38. We noted that throughout the follow-up period, children who had a higher number of GI pathogens were more stunted than those with a lower number of GI pathogens. These changes may be due to environmental enteropathy, a poorly understood, chronic condition associated with enteropathogens, gut inflammation and a leaky gut seen in children from developing countries39. Diarrhoeal pathogens in the MAL-ED study, specifically EAEC, Campylobacter, and Shigella, were associated with a reduction in linear growth (LAZ) after three months38. Additionally, 95% of stool specimens in the MAL-ED study, from predominantly non-diarrhoeal episodes, detected at least one enteropathogen – which, if persistently present, can lead to altered growth13,40.
The linkage between respiratory infection and growth is not well explored. In the Pneumonia Etiology Research for Child Health (PERCH) study, multiple respiratory pathogens were noted among children hospitalised with severe pneumonia, with an average of 3.8 (SD 1.5) pathogens/participant and 3·6 (SD 1·5) pathogens/participant among age and sex-matched controls41 which is comparable to the average number of pathogens found in our study. Additionally, higher numbers of respiratory pathogens were consistently found in malnourished children, suggesting a possible link between respiratory pathogens and growth42,43. Our participants generally did not have respiratory symptoms, and we did not see an association between prevalence of respiratory pathogens with short-term growth. Colonisation is unlikely to affect short-term growth41,44,45.
It is however worth noting that the complex interaction between nutrition, infection and immunity puts children at risk of persistent and recurrent colonisation and subsequent infections including those of the respiratory tract16–19,44. There was a high prevalence of stunting in this study population. At enrolment, over a third of our study population showed stunted growth and the mean LAZ was -1.8 (1.4)23, similar to the national stunting prevalence amongst children <24 months46. There was a significant positive change in WAZ over the follow-up period, which would be explained by catch-up growth achieved from reduced incidence of diarrhoea/infection over the follow-up period11,12,14. We did not see any change in WLZ and WAZ. The majority of samples from this population that had a high number of stool pathogens also had a high number of pathogens in sputum. While an unhealthy gut microbiome composition is thought to be a risk factor for respiratory infections16,19, we cannot make any inferences or conclusions from these data.
Our study has limitations. The sample size was small, and not powered to detect changes in short-term growth. This was a secondary analysis, and we did not have a comparison group of children with no diarrhoea/Cryptosporidium spp. infection at baseline. Stool or respiratory samples collected at the time of recruitment were not evaluated for pathogens beyond Cryptosporidium, and therefore we could not assess change in pathogens from enrolment. Although we collected HIV status, data were not available for 84% of the participants. However, the strengths of our study include the serial sampling, clinical, and anthropometry data collected over an eight-week period, the collection of induced sputum from the respiratory tract, and the high sensitivity and specificity of the molecular methods we used26,45.
In summary, among young children hospitalised with diarrhoea, multiple gut and respiratory pathogens were prevalent in the participants over the following eight weeks, and the presence of more GI pathogens, but not respiratory pathogens, was associated with reduced short-term growth. Further study of larger cohorts is warranted, to delineate how gut and respiratory pathogens interact and contribute to linear deficits, during a period where insults that occur can impact long-term growth, developmental, and cognitive outcomes26.
Written informed consent for publication of the participants’ details was obtained from the participants.
Figshare: Recruitment and follow-up data of participants recruited in the sub-analysis of the CryptoResp study, https://doi.org/10.6084/m9.figshare.2126614247
This project contains the following underlying data:
Data file 1: A dataset of participants presenting with diarrhoea and followed up over an 8-week period at Queen Elizabeth Central Hospital.
Figshare: CONSORT checklist and flow chart for ‘Respiratory and diarrhoeal pathogens in Malawian children hospitalised with diarrhoea and association with short-term growth’ https://doi.org/10.6084/m9.figshare.2126857248
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We thank the patients and parents for participating in this study. We thank the research clinical and laboratory staff for conducting the study. We thank David Moore and Tanja Adams with training the CryptoResp clinical and laboratory teams. We thank Neema Toto for interim study support, Thokozani Ganiza for assistance with data management, and Wes Van Voorhis for reviewing a draft of the manuscript and providing critical feedback.
| Views | Downloads | |
|---|---|---|
| Gates Open Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Diarrheal disease, biostatistics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: I am physician-scientist. My clinical research involves diarrheal diseases, malnutrition, sepsis and critical illnesses.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 22 Nov 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with Gates Open Research
Already registered? Sign in
If you are a previous or current Gates grant holder, sign up for information about developments, publishing and publications from Gates Open Research.
We'll keep you updated on any major new updates to Gates Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)