Keywords
time series analysis, program evaluation, vaccine evaluation, power calculation
time series analysis, program evaluation, vaccine evaluation, power calculation
After a new vaccine is introduced, it is often necessary to evaluate the effect of the intervention on disease rates. This is typically done by evaluating changes in the average number of cases or the trend in cases before and after vaccine introduction1. However, this type of analysis is challenging because it can be difficult to distinguish changes in disease rates caused by the vaccine from changes resulting from random variations, epidemics, changes in healthcare utilization, or changes in reporting practices. Additionally, there is often no ‘ground-truth’ against which estimates can be compared to determine whether an estimate is credible. These issues are a threat to the validity of any vaccine evaluation study, even when using large nationwide databases. However, the problems are especially acute when moving from larger to smaller populations, where the signal to noise ratio will be lower.
While it is intuitive that having more noise in the data makes it more difficult to detect a change, it is not clear how much data are needed to effectively quantify a vaccine-associated change if one exists. The power to detect a decline will depend on many factors, including the magnitude of the expected effect (higher power with a greater expected decline), the number of cases per unit time, and the number of years of pre- and post-vaccine data. Because the specific characteristics of datasets can vary, it is difficult to make general statements about power. However, simulation-based methods can be used to evaluate and compare power in different datasets based on the pre-vaccine time series and the magnitude of the expected effects2,3.
In this study, we present a simple web-based tool that can be used to input any disease time series and obtain an estimate of the power for that series to detect a specified vaccine-associated decline. This is accomplished by extracting characteristics of the time series (e.g., seasonality, trends, unexplained variability) from the pre-vaccine period and simulating a set of time series that have similar characteristics and have a vaccine impact that is specified a priori. We demonstrate the application of this approach on observed pre-vaccine data on pneumonia hospitalizations from the 26 states plus the federal district in Brazil and relate characteristics of the time series to the power.
We used state-level hospitalization data from Brazil, which have been described in detail previously4. These de-identified data are drawn from the Unified Health System (SIH-SUS, Ministry of Health), which captures ~70% of the population in Brazil. The raw data can be obtained directly by contacting the Ministry of Health in Brazil. The formatted time series data are available in the Github repository for this study. Each hospitalization is assigned a unique ICD10 code. For these analyses, we focused on data on <12 month old children and 80+ year old adults for the pre-vaccine period 2003–2009. These two populations provide a useful contrast. The time series for the infants was relatively stable prior to vaccine introduction, while the data for the 80+ year old adults had a notable increasing trend before vaccine introduction. Both sets of time series exhibit strong seasonality with a peak in the winter.
The goal for this exercise was to simulate a set of time series with characteristics that resembled the observed hospitalization data from the pre-vaccine period but that had specified vaccine effects added in. We then sought to estimate the vaccine effect using the same model that was used to generate the data and evaluate the power to detect the effect. This provides a best-case scenario where the underlying model is correctly specified.
The first step in this process was to extract characteristics of the time series from the pre-vaccine period (trend, seasonality, and amount of unexplained variation). For each state, we fit a regression model to the data from the pre-vaccine period (2003–2009). The outcome variable was the number of pneumonia hospitalizations (coded as J12-18) per month, and the covariates were an index variable for time (to capture any linear trends in the data) and 12-month and 6-month harmonic variables (to capture seasonality). We used a Poisson regression model with a Gaussian observation-level random intercept to account for overdispersion in the data such that
The models were fit using the glmer function in the lme4 package in R, version 3.6.1.
The next step is to use the fitted model to simulate time series of counts of hospitalization with similar characteristics that covered both the pre- and post-vaccine period, with a specified vaccine-associated decline added to the simulated data. Using the estimated regression coefficients and their estimated variance/covariance matrix, we generated 500 independent random draws of the parameters from a multivariate normal distribution for each state and age group combination. These were combined with the design matrix to obtain simulated Random draws of were independently generated from a normal distribution with a mean of 0 and a standard deviation equal to the standard deviation estimated for ϕt from the fitted model. To incorporate a known vaccine effect, we assumed that the time series declined by a specified amount over a 24-month period. We generated time series where the maximum vaccine effect after 24 months ranged from a 10% – 50% reduction (rate ratio of 0.5–0.9). To capture these declines, we generated a vector, vt, with entries equal to 0 prior to vaccine introduction and decreasing linearly to the log(Rate-Ratio-Final) over 24 months. Simulated counts, Yt,sim, were generated by taking a random draw from the Poisson distribution with mean The simulated counts reflect uncertainty in the regression parameters, unexplained variability in the data, as well as uncertainty from the observation process.
We next used a regression model similar to the one used to generate the data to estimate the vaccine effect. The outcome was the (simulated) number of counts per month. As above, we adjusted for seasonality using 6- and 12-month harmonic terms, and secular trends were captured using an index for time. The vaccine effect was quantified using a linear spline term that began at the time of vaccine introduction and continued for 24 months before stabilizing. An observation-level random intercept was included to capture overdispersion of the count data. Using the fitted model, we calculated the estimated rate ratio 24 months after vaccine introduction as 24*(coefficient for the vaccine effect term). To evaluate how many years of pre-vaccine data are needed to estimate the effects, we sequentially removed the first 1, 2, or 3 years of data and evaluated the effect on power.
All of the time series data and code used in these analyses are available from a Github repository https://github.com/weinbergerlab/PoissonITS_power. The interactive tool, along with a sample dataset, can be accessed at https://weinbergerlab.shinyapps.io/ITS_Poisson_Power.
Because the power to detect a change in a time series is influenced by the expected effect size, the amount of unexplained variation in the data, and the number of years of data available, it can be difficult to make general statements about power. However, observed time series from the pre-vaccine period can be used to simulate time series to perform a best-case power calculation. This can provide an indication of whether it is worth performing an analysis or whether collecting additional data (e.g., additional pre-vaccine time points) could be helpful. We provide a simple ‘point-and-click’ interface where the user provides a time series in a csv or Excel format, indicates which columns contain the date variable, the outcome, and any potential controls, and the date at which the intervention is introduced (Figure 1).
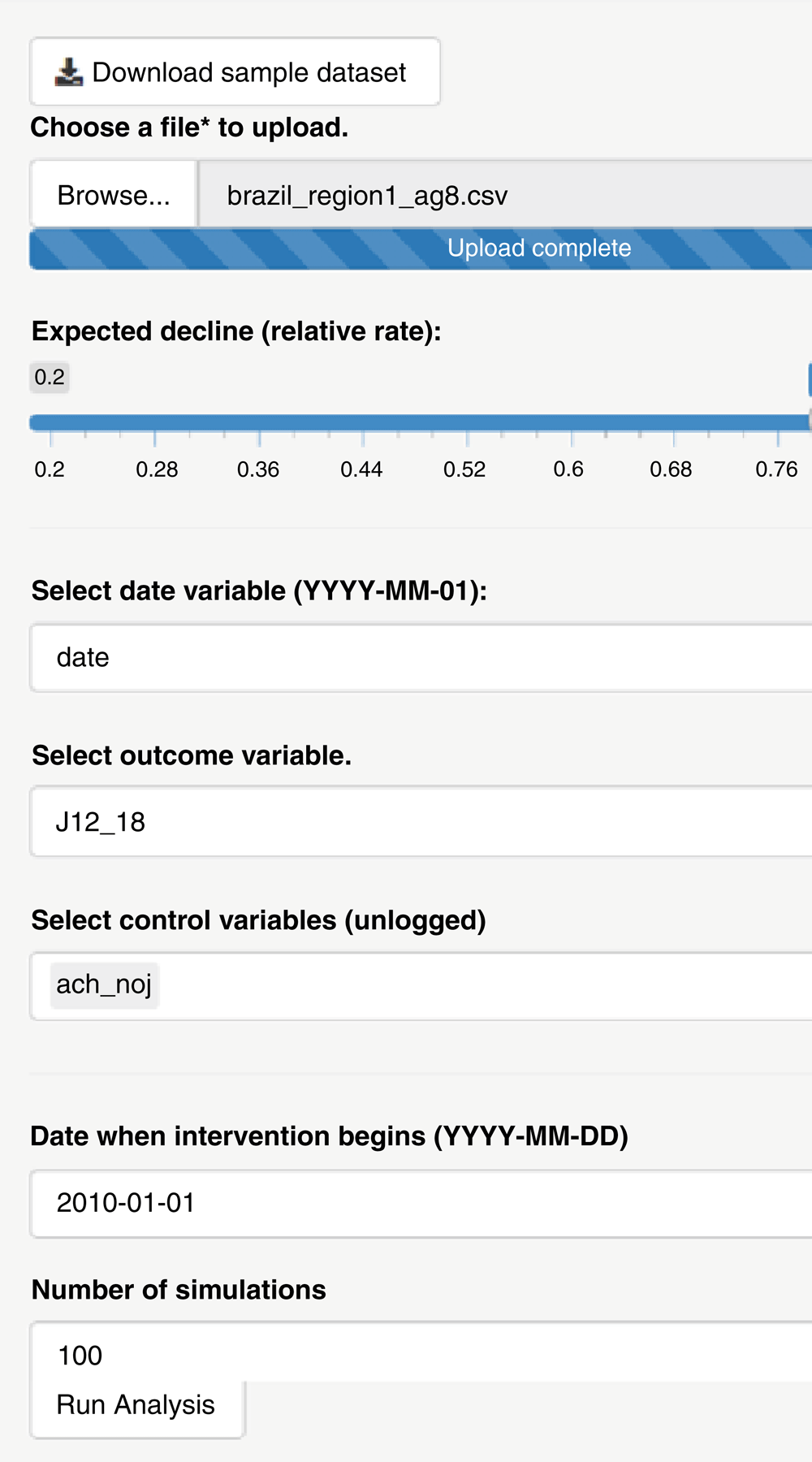
The user uploads a time series, specifies the expected decline in terms of a rate ratio, specifies the key variables (date, outcome of interest, and controls), the date of the intervention, and the number of simulations to generate. A sample dataset can be downloaded by clicking the button at the top of the screen.
As a demonstration of this approach, we apply this simulation framework to data from Brazil, disaggregated to different subnational levels (state, region). The size of the population varies drastically by state, from 450,000 to 41 million individuals (in 2010). On average there were 30-1900 hospitalizations due to pneumonia per month per state among children <12 m and 12-1100 hospitalizations per month per state among adults 80+ years of age during the pre-vaccine period. The time series for the <12m old children were highly seasonal but without a strong long-term trend, while the time series for the 80+ year olds increased markedly starting in the pre-vaccine period. We simulated time series for each of the states that had similar characteristics to the observed time series in the pre-vaccine period but with vaccine effects of different magnitudes (Figure 2).
We first evaluate the relationship between the amount of unexplained variability in the data and the ability to accurately estimate the effect of the vaccine. There is a clear relationship between the amount of unexplained variability in the data and the power to detect a vaccine-associated change (Figure 3A). This trend was consistent across all of the states between both children and adults.
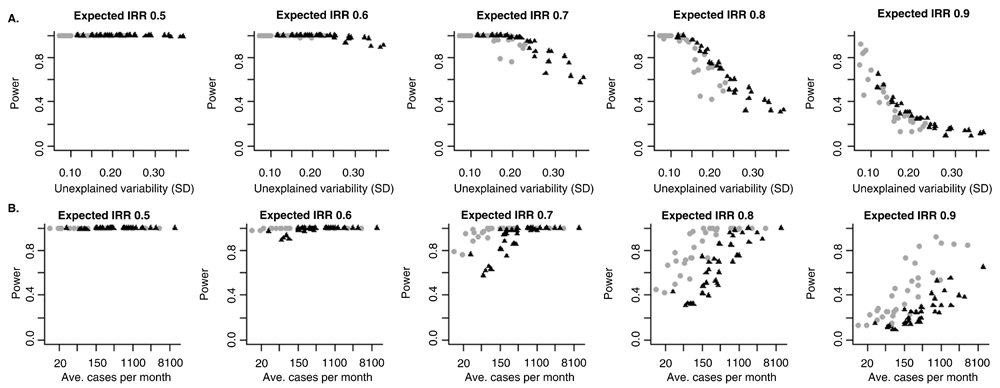
Relationship between power to detect a decline associated with vaccine introduction and (A) the amount of unexplained variation in the time series or (B) the average number of cases per month for different specified magnitudes of vaccine effects. The labels at the top of the panel indicate the magnitude of the expected vaccine effect, with an incidence rate ratio (IRR) of 0.5 representing a 50% decline associated with the vaccine and a IRR of 0.9 equal to a 10% decline. Each dot represents the power for one state in Brazil. The black triangles represent estimates for adults 80+ years of age, and the gray circles represent estimates for children <12 months of age.
Plotting the estimated power against the average number of hospitalizations in the state/region, there is also a relationship, but the trend differed between children and adults (Figure 3B). This is because the amount of unexplained variability was higher in the <12m old children than in the 80+ year old adults (Extended data: Figure S1)
With fewer years of baseline data, the power to detect a change in disease rates associated with the vaccine also declines. For datasets with little unexplained variability, even with just 12 months of pre-vaccine data, there could be high power to detect a vaccine-associated decline of 20%. However, when there is more unexplained variability in the time series, power declines with shorter pre-vaccine periods (Figure 4). These declines in power are particularly dramatic for time series with intermediate levels of unexplained variability (Figure 4).
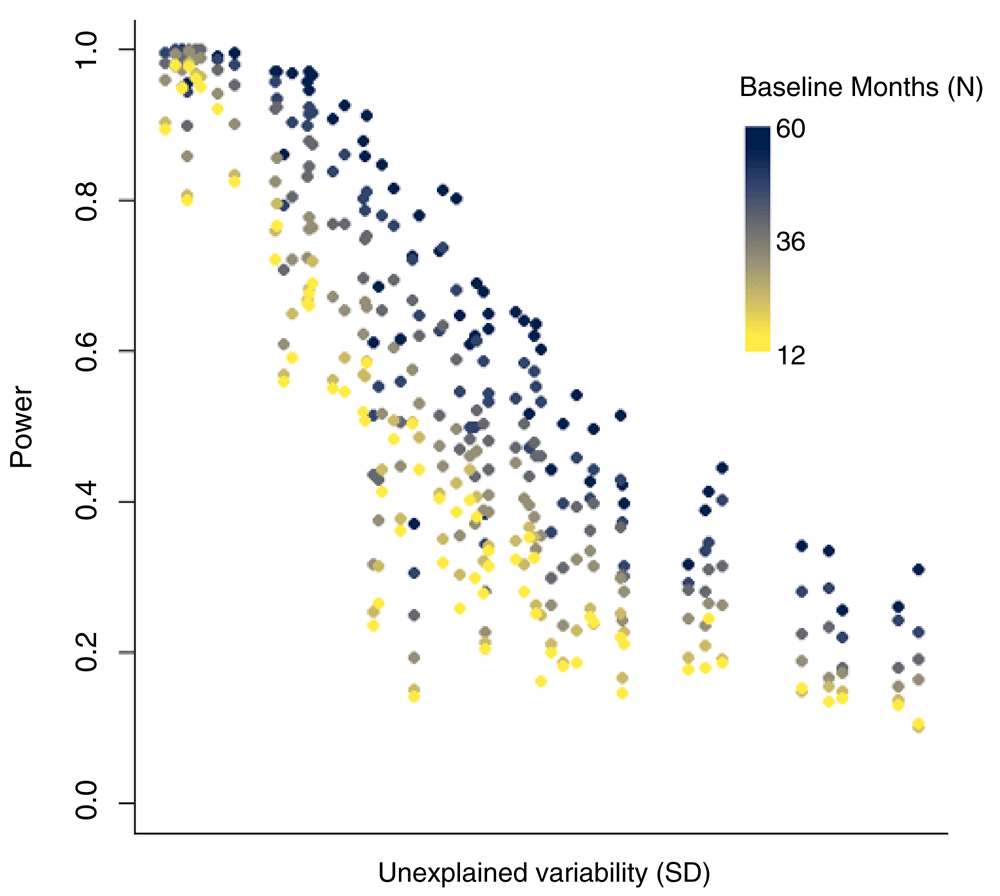
Each dot represents the power for one state/age group in Brazil. Dots with lighter colors had fewer years of data.
As a demonstration of the point-and-click interface, we use hospitalization data from Chile among children <24 months (raw data available from http://www.deis.cl/)5. This sample time series can be downloaded directly from the interface. The outcome variable is the number of hospitalizations per month due to all-cause pneumonia (J12_18) for 2003–2014. The number of non-respiratory hospitalizations per month (ach_noj) is included as a control. If no control is present, this field can be left blank. The date of vaccine introduction is set to January 1, 2011. The program generates a specified number of simulated time series (N) based on the pre-intervention data (Figure 5A). With three years of post-vaccine data (vaccine introduction in 2011, evaluating through 2014), the 100 estimates of the rate ratio are centered on the true value (indicated by a red dashed line), with a moderate degree of uncertainty (Figure 5B). This yields 54% power to detect a rate ratio of 0.8 (Figure 5B). Compared with the analyses of the Brazil series, the power is reasonable given the amount of unexplained variability in the data but could be increased (Figure 5C). This can be seen in the simulation by increasing the length of the evaluation period by a year (e.g., in this instance moving the date of vaccine introduction earlier by 12 months to January 2010, which also shortens the baseline period) (Extended data: Figure S2). The power in this instance would increase from 54% to 77% with an improvement in the precision of the estimates. This also highlights that the total number of years of data is not the only key component—it is how that data points are balanced between the pre- and post-intervention periods—too few data on either side will reduce power.
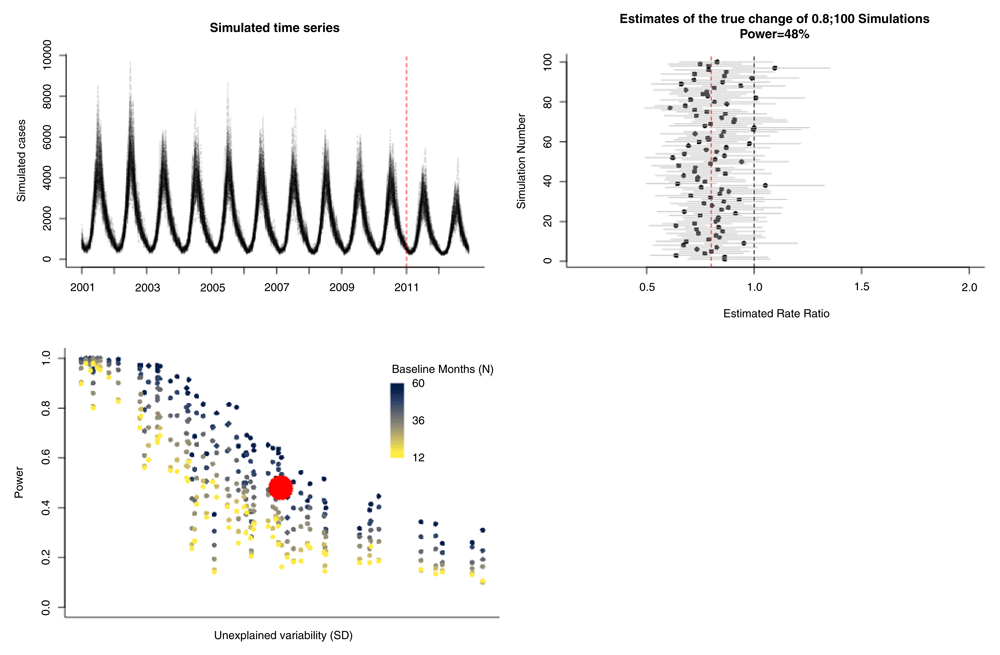
The upper left panel shows the 100 simulated time series. The upper right panel shows the estimates of the rate ratio for each of the 100 simulations. The true specified rate ratio (0.8) is denoted by a red dashed line. 54% of the estimates had 95% confidence intervals that did not cross 1. The bottom left panel shows the estimate of power for this study (red dot) compared with the estimates from the Brazil states with different length baseline periods.
In this study, we describe a simple interface for conducting simulations to evaluate the power to detect a vaccine-associated decline from time series data. This approach provides analysts a simple best-case scenario for determining whether they are likely to detect specified vaccine effects with the data on hand or whether collecting additional pre- or post- vaccine data would be beneficial. This type of tool should be used when planning analyses and prior to conducting a formal evaluation analysis with the data on hand.
By analyzing subnational data from Brazil, we demonstrate how power varies with the number of cases and the degree of unexplained variability in the data. Reducing unexplained variability in the data by using time-varying covariates can help to increase power. Such covariate could include other causes of disease/hospitalization/death or known correlates of changes in disease rates (e.g., percent of the population with access to healthcare).
These analyses evaluate power based on the statistical characteristics of the time series. As with any analysis, failure to correctly control for relevant trends will also introduce important biases and could greatly outweigh the issues related to statistical characteristics of the data. For instance, if there is a non-linear trend that is not well-captured by an interrupted time series analysis, the vaccine effect could be substantially over- or under-estimated.
The estimates generated with this approach represent a ‘best-case’ scenario where we know the exact date of vaccine introduction and where all non-vaccine-associated changes are linear and can be controlled with a simple model. In reality, numerous factors can influence pneumonia hospitalization rates. The use of control variables can help to adjust for these, but often remain unexplained factors that cannot be easily adjusted.
We summarize the results of these simulations in terms of statistical power (i.e., what percentage of simulations yielded a statistically significant effect when an actual non-zero effect was present). In practice, we typically avoid describing evaluations of vaccine impact made using observational time series data in terms of statistical significance. It is often more informative to instead describe the estimate of vaccine impact and the strength of the evidence/precision of the estimates. These types of analyses are rarely used for making dichotomous policy decision (e.g., licensure), so using an arbitrary threshold for declaring whether a vaccine ‘works’ is not needed.
In conclusion, we present a simple framework for evaluating the power to detect vaccine-associated declines of a specified magnitude. This approach can help in planning for an evaluation study and for understanding differences between studies.
The Brazilian dataset can be accessed by contacting the Ministry of Health (Ministério da Saúde) directly via http://portalms.saude.gov.br.
Chilean dataset can be accessed from the Chilean Department of Statistics website: http://www.deis.cl
Time series data and code available from: https://github.com/weinbergerlab/PoissonITS_power
Archived data and code as at time of publication: http://doi.org/10.5281/zenodo.36897316
License: CC0
Figshare: Extended Data Figure S1, https://doi.org/10.6084/m9.figshare.119081437
Figshare: Extended Data Figure S2, https://doi.org/10.6084/m9.figshare.11908158.v28
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Interactive tool available from: https://weinbergerlab.shinyapps.io/ITS_Poisson_Power
Source code available from: https://github.com/weinbergerlab/PoissonITS_power
Archived source code as at time of publication: http://doi.org/10.5281/zenodo.36897316
License: CC0
| Views | Downloads | |
|---|---|---|
| Gates Open Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Pediatric infectious disease epidemiology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Hyndman RJ, Athanasopoulos G: Forecasting: principles and practice. OTexts. 2018. Reference SourceCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 19 Oct 20 |
read | |
|
Version 1 09 Mar 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with Gates Open Research
Already registered? Sign in
If you are a previous or current Gates grant holder, sign up for information about developments, publishing and publications from Gates Open Research.
We'll keep you updated on any major new updates to Gates Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)