Keywords
mortality data, reporting delay, real-time, nowcasting, epidemiological modeling, smoothing
This article is included in the Coronavirus (COVID-19) collection.
mortality data, reporting delay, real-time, nowcasting, epidemiological modeling, smoothing
What policy makers and analysts are interested in during an outbreak is the times series M of number of deaths mt that happened on each day t,
During the outbreak, this is typically not what is available in real time. Instead, we shall have a closer look at two commonly used times series. The first time series is what has been reported by countries and regions to the WHO1, the US CDC2, the European CDC3 and that eventually end up in dashboards like Johns Hopkins University4,5 or Our World in Data6. This is the number of new deaths rt that has been reported to competent agencies in countries and regions during the last 24 hours, and which the countries and regions in turn report to the WHO and the European CDC on the reporting date t. We shall call this the reporting series, R,
Note that this number is only on very rare occasions updated retrospectively. Normally, a new number is added to the time series every day. Also note that we know nothing about when the deaths happened, the event date, we only know the number of deaths reported on each day.
The second time series is a time series that some countries, e.g. Sweden7–9, Belgium10 and the UK11,12, make available on the internet. This data is normally a de-identified extract from the national surveillance and reporting system. This series has information about when the deaths actually happened. We will call this data set the event-based series DT,
This data set is a time series reported on day T, where each number in the series, is the cumulative number of deaths on each event date t known to the country agency on date T. Note that typically a whole new time series is reported every day. As time goes by, more deaths find their way through the reporting process, the number of deaths for every event date in the past is updated.
The two time-series as reported by Sweden on 2020-05-01 have been plotted in Figure 1. A few general comments about these curves. The R-series varies dramatically with a weekly pattern, whereas the DT-series varies less but descends sharply towards zero as time approaches the reporting time T. From inspecting the cumulative versions in Figure 2 one can see the R-series for the most part represents a delayed situation compared to the DT-series, but that they coincide on the reporting day. Notably, these times series do look quite different and in what follows we will discuss how to interpret these time series and where care must be taken in using these data sets as input in modelling or for decision making.
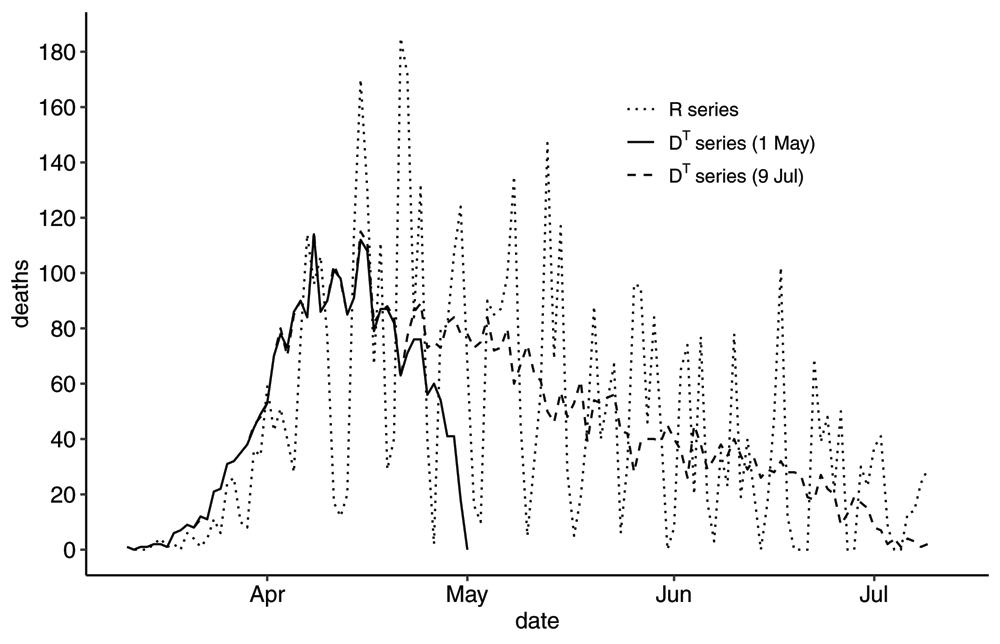
he R-series and the DT-series as reported by Sweden on 2020-05-01 and on 2020-07-09. It is enough to plot just one R-series as it does not change retrospectively unlike the DT-series, which is updated retrospectively every time it is reported.
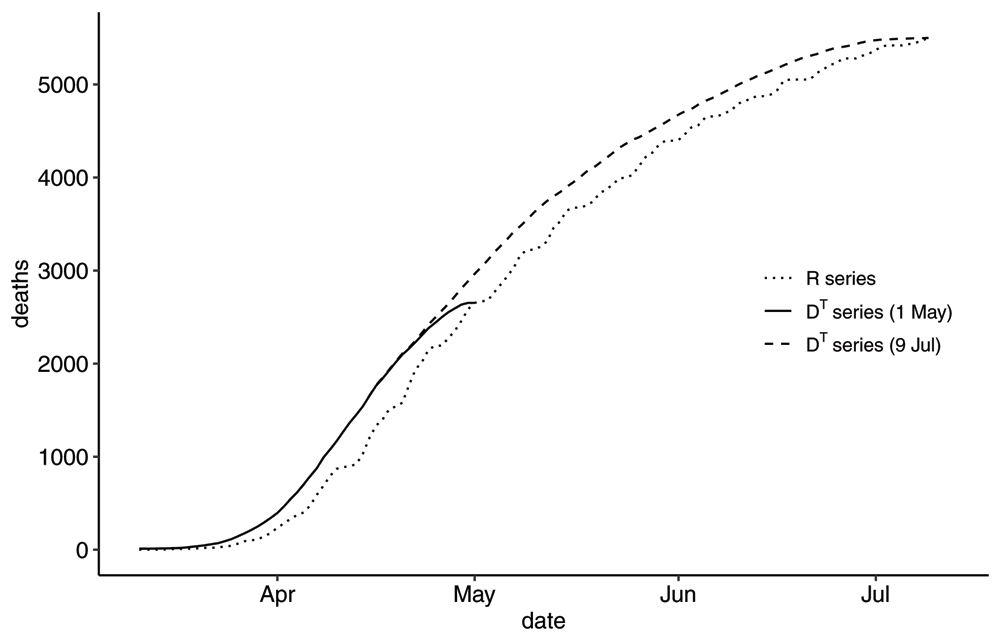
The cumulative R-series and the cumulative DT-series as reported by Sweden on 2020-05-01 and on 2020-07-09. Clearly, there is a delay in the R-series compared to the DT-series. The cumulative number of deaths coincide for both series on the reporting day.
In addition to comparing the R-series and the DT-series, we will say a few words about nowcasting, and introduce a naive method based on the characteristics of the reporting process.
It turns out that there is a rather elegant mathematical framework for the two series in terms of matrix algebra. For the interested reader we have put all of that in the Appendix (see Extended data13). One of the key findings is equation (A16) which is the mathematical relationship between the R-series and the DT-series which is worth repeating here. The relationship between rt and is
where is the so-called forward-looking probability mass function (pmf) defined as the fraction of the deaths that happened on event date e and were reported exactly Δ days later. If there were underlying stochastic variables for when deaths happened and were reported, would have the interpretation as an estimate of the conditional probability P(reporting date = e+Δ | event date = e). There is also a corresponding forward-looking cumulative distribution function (cdf) defined as the the fraction of the deaths that happened on event date e and were reported within Δ days.
The expression (4) makes intuitive sense. It says that the number of deaths reported on day t, is the weighted sum of deaths happening on previous event days. The contribution from each event day e is the total number of deaths that day times the probability that those deaths will be reported exactly t–e days later.
Now, typically the reporting process does not change that much over time, in which case one may want to find the average cumulative distribution function and the average probability mass function. See expression (A15) in the Appendix (see Extended data13) for how this is done. Also see the equation (A20) in the Appendix (see Extended data13) for the inverse of (4), i.e. as a function of rt.
Equation (4) is interesting from another perspective. It is the defining expression of a linear time-dependent filter, where in-signal x(t) is modulated by a time dependent filter function f (τ, t, –τ) resulting in an out-signal y(t)
In mathematical terms, the expression (5) is a so-called convolution of the true signal and the time-dependent filter. Filters are used in many areas including epidemiology, although its main application is in signal processing. Interestingly, studies of time-dependent filters relevant to this paper can be found in geology, e.g. in14,15. The main insight here is that the practice of reporting daily deaths, the R-series, as is standard procedure, should be viewed as a time-dependent filter modulating the event-based curve. The reporting series R is distinct from the true death curve, even if it in most practical applications comes very close. Below we will highlight some circumstances where this difference can be important.
As can be seen in Figure 1, the DT-series typically descends to a value close to zero on the reporting day T. This is because few deaths will be reported on the same day they actually happened. Many deaths have happened but are still being processed in the reporting system and will be reported at a later date. This means that for a time period prior to the reporting time, the DT-series will underestimate the actual number of deaths on those days. Ideally, one would like to know the final number of deaths on any particular event date e before the reporting date T, i.e. finding the M series for t < T. This is the problem of nowcasting. Several groups have devised algorithms to estimate the M-series for t < T, see16–22. In the Appendix (see Extended data13) we derive a very simple expression for the M-series, i.e. for me = , which is
Intuitively this expression makes sense. It says that one could get the final number of deaths on an event day e by dividing the number of deaths on that day that have been reported so far (i.e. ) by , which the fraction of the final number of deaths on day e reported within T–e days since day e. Of course, at time T we don’t know what the final cdf will look like but by making the assumption that the reporting process will behave as it has done in the recent past, we will be able to compute simple nowcasts below. The drawback of this method is that it does not work when , but as we will see it can still be quite useful.
The computations in this paper have been based on the fundamental data set which was constructed from downloading the DT-series daily from 2020-04-02 to 2020-07-09. Data processing was done in R-studio (v1.2.1335) using R version 3.6.0 (2019-04-26). An R-script including relevant R-packages has been made available. Further information can be found in the Data availability section1323.
We will now apply the methods described above and, in the Appendix (see Extended data13), to analyze the relationship between the reporting series and the event-based series as reported by Sweden.
As we have argued, the reporting process is characterized by the two time-dependent distribution functions and . In Figure 3 we have plotted the cumulative distribution functions for all event dates between 2020-04-02 and 2020-06-01, as well as the resulting mean cdf.
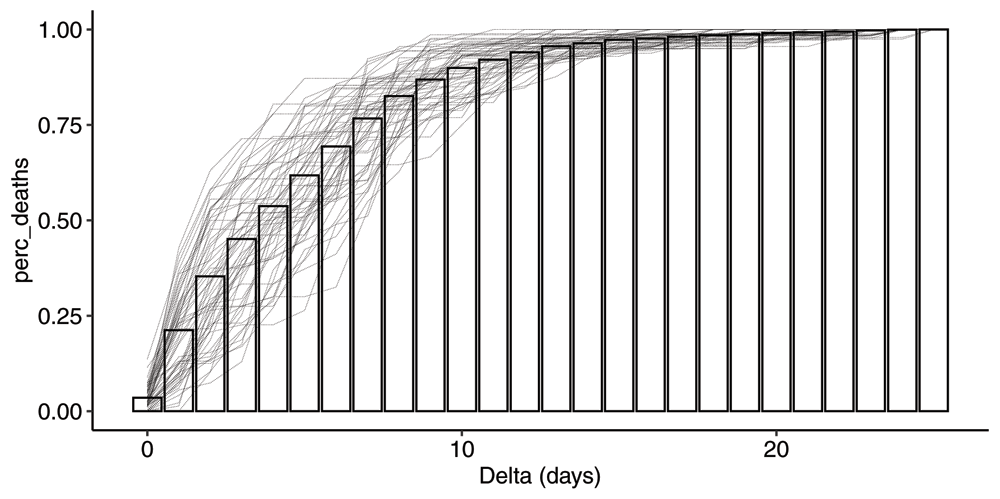
Plot of all cdfs for event dates between 2020-04-02 and 2020-06-01. The bars represent the mean cdf which is time independent. The cdf is defined as the fraction of deaths on a particular event date e that will be reported within Δ days.
In Figure 4 we show the corresponding mean pmf . The first observation is that there is a significant delay in the reporting process. The average delay can be computed using equation (A14) and is 5.2 days. By inspection of the cdf, we see that it takes on average 7 days to capture 75% of the deaths on a particular day, and about 10 days to capture 90% of the deaths.
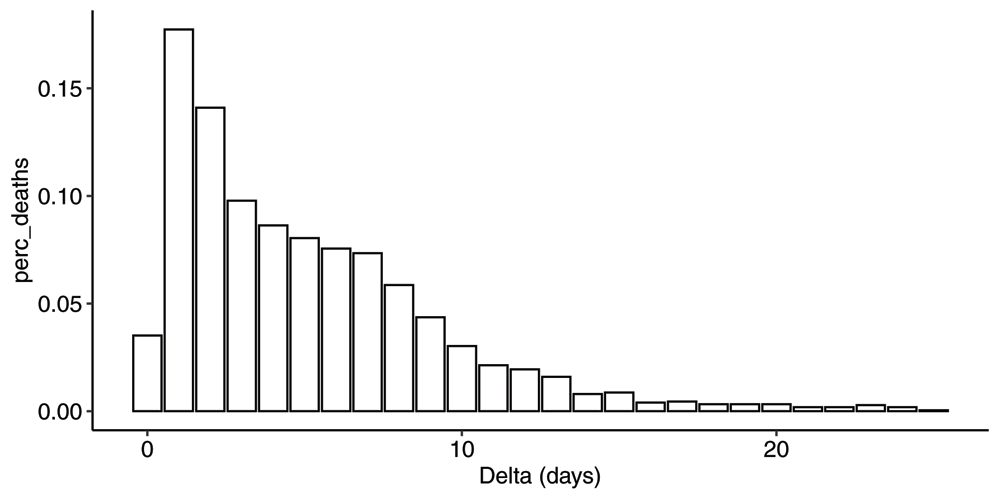
Plot of the mean probability mass function corresponding to the mean cdf across all event days between 2020-04-02 and 2020-06-01.
Furthermore, from inspection of the pmf in Figure 4, there seems to be a substructure to the reporting process. One can identify three sub-processes with different delays. Allow us to speculate that we have one reporting process for deaths in hospitals with an average delay of 1–2 days, one process for deaths in nursing homes with an average delay of 6–7 days and a third process that for some reason takes even longer, with an average delay of 11–12 days. One can imagine the pmf resulting from superposition of three distributions where the respective areas under the curve correspond to the fraction of deaths in those three settings respectively. Again, by inspection of the pmf, we estimate that roughly 3/8, 1/2 and 1/8 of the deaths happened in those three settings respectively. This is roughly in line with what has been reported by Sweden24.
In order to better understand the weekly periodic pattern in the R-series, we have computed the average distributions for each weekday. The average cumulative distribution functions for each weekday is plotted in Figure 5. There is a flat portion of each curve representing the fact that deaths were essentially not reported on Sundays. For Saturdays the flat portion is between day 0 and day 1; for Fridays between day 1 and 2 etc. Sweden stopped reporting both the R and the DT-series on weekends from 2020-06-20 onwards.
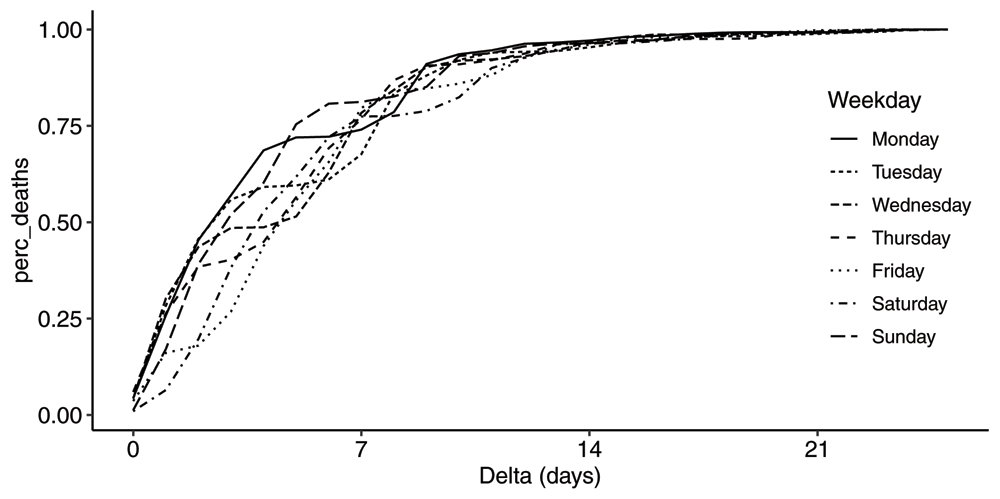
Plot of the average cdfs by weekday, for all event days between 2020-04-02 and 2020-06-01.
We also plot the pmfs in Figure 6. Given the simplicity of the mean distribution, the differences by weekday are a little surprising. It is a complex interplay between the three sub-processes mentioned previously and a significantly reduced reporting on Saturdays and Sundays resulting in the "valley" on day 6 on Mondays moving one day to the left every day.
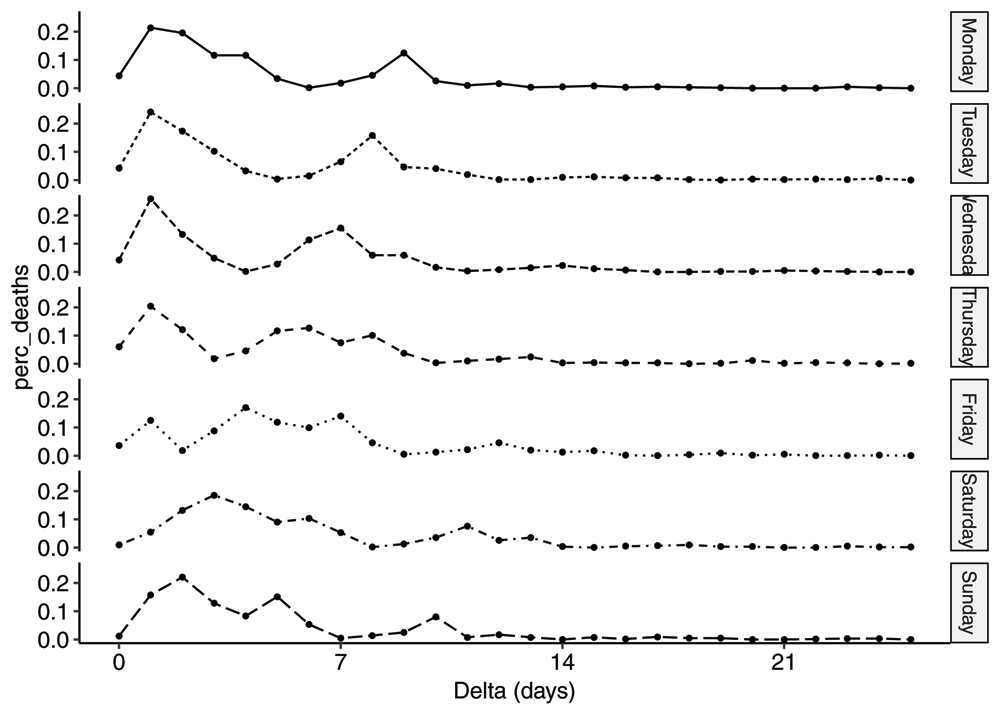
Plot of the average pmfs by weekday, for all event days between 2020-04-02 and 2020-06-01.
Next, in order to isolate the effect of the reporting process, we compute the effect of the Swedish reporting process on a hypothetical smooth bell shaped death curve, much like we have done for two simple examples in Figure A11 and Figure A12 in the Appendix (see Extended data13). The result can be seen in Figure 7.
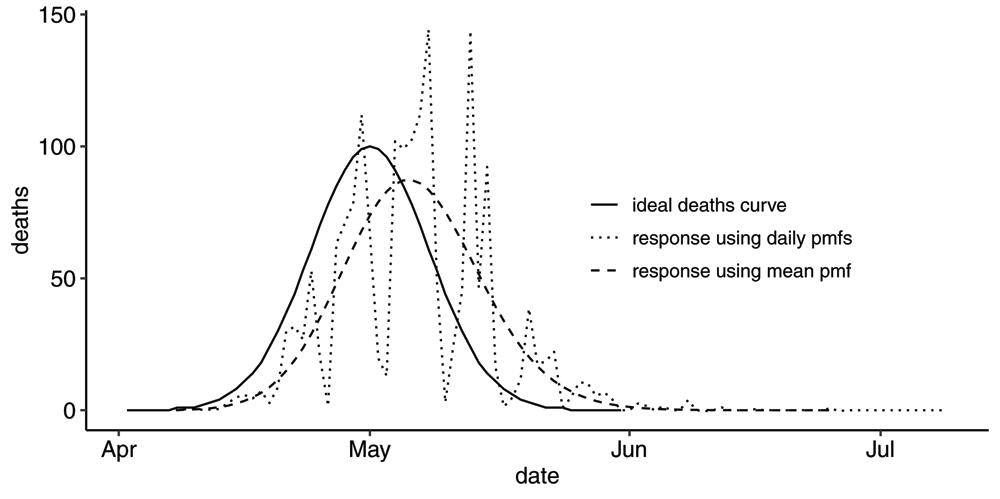
Plot of the Swedish reporting process applied to a hypothetical smooth bell-shaped actual deaths curve. The R-series resulting from application of the full time-dependent set of pmfs shows a response very similar to reported R-series. The R-series resulting from the application of the time-independent mean pmf produces a modified bell-shaped R-series which is flatter and wider with a slightly different slope.
Again, the daily, time-dependent pmfs produce a highly variable output just like the observed reporting series. Contrasting this output to the output one gets using the mean time-independent pmf, we conclude that the periodic pattern in the reporting series has its origin in characteristics of the reporting process rather than in the characteristics of the actual deaths curve. Although this is perhaps not very surprising, the amplitude of the periodic pattern is surprising. Relatively small differences in the reporting processes from one weekday to the next results in these wild swings in the reported daily deaths. Is there an opportunity to give guidance to the countries for how to reduce these resulting swings? We also note that the Swedish reporting process modifies the shape of the hypothetical death curve in three ways, just like the hypothetical cases in the Appendix (see Extended data13). First, there is a clear time shift. Second, the peak is lower. Third, the slope is flatter, both as deaths increase and decrease.
As we have seen, the Swedish R-series is very variable. Hence, when using the R-series as model input one may have to do some pre-processing. If used in a time dependent model together with case incidence or prevalence (e.g. in an SIR model) one may have to adjust for the reporting delay, since the reporting of cases normally is quicker than the reporting of deaths. The average reporting delay for cases in Sweden is approximately 1–2 days.
Additionally, if initial conditions need to be specified for the magnitude as well as the slope of the death curve, some kind of smoothing of the R-series is appropriate. The question then arises, what is a good smoothing of the R-series? Since the R-series is not the same as the DT-series, what should a good smoothing of the R-series look like? The example in the previous section gives us a clue. Based on the graphs plotted in Figure 7, a measure of how good a smoothing is to see how closely it matches the "transformed" DT-series, i.e. the series one would obtain by applying a "smoothed reporting process" to the event based series, i.e. using the time-independent mean pmf rather than the time-dependent pmfs. In addition, a good smoothing should have the same area under the curve as the R-series. Unfortunately, neither the DT-series, nor the mean pmf are available to the modelers.
One model that relies on the shape of the death curve is the model developed by the Institute of Health Metrics and Evaluations (IHME) at University of Washington25. In Figure 8 we have plotted the Swedish R-series as reported on 2020-06-03 well as the smoothing of the R-series by IHME modelers in the 2020-06-05 update of their model26. We have also plotted the transformed DT-series one obtains by filtering the DT series using the mean pmf.
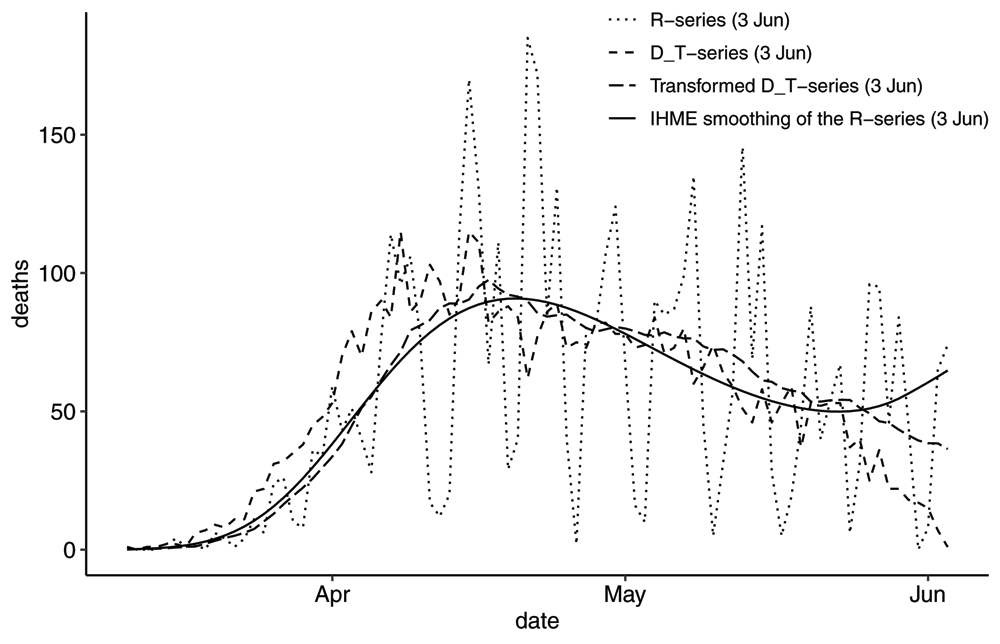
Plot showing the IHME smoothing of the R-series on 2020-06-03 in comparison with the Transformed DT-series using the mean pmf. Note there is an uptick in the R-series just before the reporting date, which may have influenced the smoothing. Also note that the Transformed DT-series has a slightly lower peak, a significant delay and a lesser slope than the DT-series.
At this particular instance the IHME modelers were unfortunate to produce a smoothing that drastically changed the outcome of the model and we can see that it deviates significantly from the transformed DT-series. This IHME smoothing preserved the number of deaths. For reference, the 2020-06-05 update of the model estimated 8357 (7046, 10386) number of deaths by 2020-08-01. A previous update on 2020-05-29, with a different smoothing, estimated 5254 (4688, 6420) number of deaths. In their next update of the model on 2020-06-25, the smoothing is very similar to the transformed DT-series.
Please note that by only using the smoothed R-series as input, one does not compensate for the delay in the reporting process, nor for the change in shape of the curve. This means for the total number of deaths, as well as the rise (or decline) in number of deaths is underestimated.
Turning our attention to the DT-series, we were curious to see how a state of the art nowcasting algorithm would perform in the presence of the strong weekly patterns in the Swedish time-dependent reporting process. We have therefore compared a "naive" approximation of the nowcasting formula (6) with the nowcasting algorithm developed by a group at Harvard17. We wanted to use both a time independent and a time-dependent approximation and used the following two naive approximations
where is the mean cdf computed as an average of the 14 cdfs for the event days 21–34 days before the reporting time T. Note that we have to go 21 days back in order to have the cdf defined for Δ between 0 and 21 days. We deliberately chose a multiple of 7 to match with the correct weekday. Starting on Sunday 2020-05-10, we have plotted 7 nowcasts of the DT-series. The first graphs are shown in Figure 9 and the following six in Figure 10.
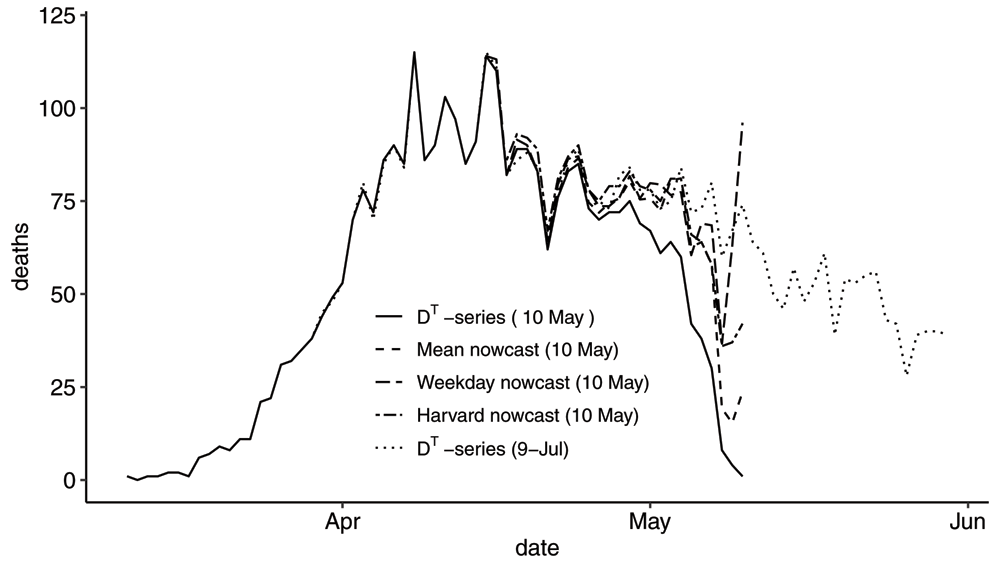
Graph showing the DT-series, the Mean nowcast, the Weekday nowcast and the Harvard nowcast of the DT-series on the reporting day 2020-05-10. The DT-series as of 2020-07-09 serves as the true M-series. On this reporting day, the three nowcasts are quite different for the two days prior to the reporting date.
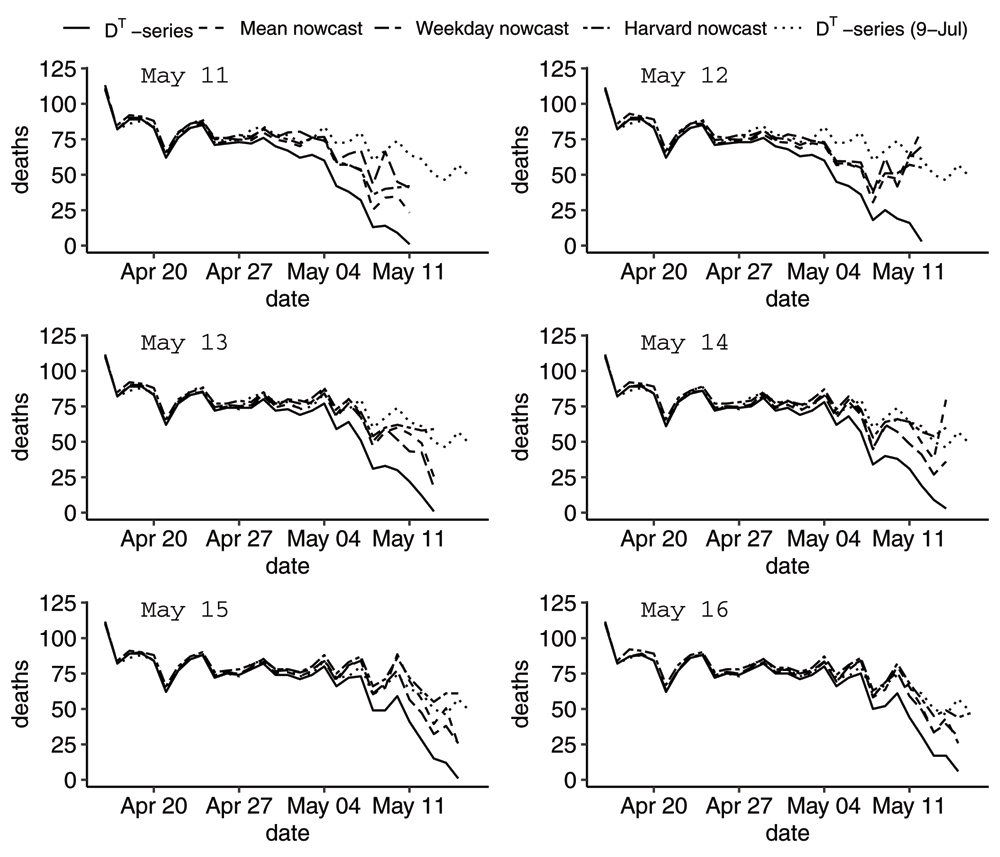
Graph showing the zoomed-in DT-series, the Mean nowcast, the Weekday nowcast and the Harvard nowcast of the DT-series on reporting days between 2020-05-11 and 2020-05-16. The DT-series as of 2020-07-09 serves as the true M-series. Although the Harvard nowcast is better, the naive nowcasts can be useful.
Generally, the Harvard nowcast algorithm performs better, but since also the Harvard nowcast fluctuates, it would be interesting to see if there are improvements that can be made by taking the weekly pattern better into account. It should be noted that we have used default settings for the Harvard nowcast. Furthermore, one can likely improve upon the approximations (7) and (8) but nowcasting is not the focus of this paper. The takeaway message here is that there are good nowcast methods available to analysts if they have access to the fundamental data set and if they are interested in using the best possible input in their analyses.
By viewing the R-series and the DT-series as two aspects of the same underlying data set, we conclude that the R-series is the result of a reporting process that "filters" the event based series, the DT-series, see Equation (4). In many cases this does not result in a significant difference between the two series. However, it turns out that the Swedish reporting process for Covid-19 deaths happens to generate a R-series that looks quite different from the DT-series. The R-series is wildly varying in time, which makes it hard to see trends and use as model input.
To remedy this for the Swedish R-series, smoothing of the curve is appropriate. Unfortunately, it is hard to know what constitutes a good smoothing, unless you have access to the DT-series. However, three factors are worth paying close attention to. First, most reporting processes have a built-in delay, leading to a corresponding shift in time between the R-series and the DT-series. Second, if the delay is significant, the shape of the curve will also be affected. The peak will be lower, and the slope of the smooth R-series will be less than the slope of the DT-series. This may result in an estimate of number of deaths that is an under-estimate during times of increasing daily deaths counts, and an over-estimate during times of decreasing death counts. Finally, if the slope of the deaths curve just before the reporting time is of importance, one should note that the perfectly smooth R-series will to some degree reflect the slope of the DT-series and drop off just before the reporting time. Getting the slope right based on the R-series can therefore be a very hard problem, as we have seen in a real world case shown in Figure 8. It is worth noting that smoothing of the R-series by applying a 7- or 14-day rolling average, which has been very common when reporting Covid-19 deaths, might be a good idea but it suffers from the same short comings as mentioned above. It adds another four (alt. eight) days of delay, dampens the peaks, and additionally flattens the slope of the death curve.
So, what is the best method? In cases where the there is a significant difference between the R-series and the DT-series, it is probably not very controversial to be of the opinion that one should use the DT-series. However, one then faces the problem of a DT-series that drops off to zero close to the reporting date, see Figure 1, and one is forced to use a nowcasting method to compensate for this drop-off. As we have seen in Figure 9 and Figure 10, the nowcasting series are not perfect and do fluctuate when applied to the Swedish data set. Nevertheless, whereas the change in shape of the R-series from one day to another can be significant, the shape of DT-series never changes that much since it distributes the new deaths over many event dates. This is the main reason why using the DT-series with nowcasting is preferable to using the R-series with smoothing, shifting and potentially compensating for a difference in shape.
Coming back to the Swedish situation, back in May it was not a trivial task to interpret the death curve and seeing the trend if you only had access to the R-series. However, having access to the DT-series and a nowcasting algorithm did give you confidence that the number of deaths were indeed declining. Of course, access to data for hospitalizations, ICU occupancy and case incidence gives a more complete picture and help interpret the situation. Nevertheless, it would be welcome if more countries made the DT-series available.
The primary data set analyzed in this paper was constructed by downloading the DT-series daily from the Public Health Agency of Sweden7 between 2020-04-02 and 2020-07-09. The data is publicly available and is considered public domain data.
Zenodo: On the use of real-time mortality data in modelling and analysis during an epidemic outbreak – underlying data
https://doi.org/10.5281/zenodo.399234523.
This project contains the following underlying data:
FHM_Covid_Download.zip. (Zip-archive of raw downloaded files with Swedish deaths data.)
swedish_covid_deaths_data.csv. (Swedish deaths data collated from the raw data files in a .csv format.)
swedish_covid_deaths_data.xlsx. (Swedish deaths data collated from the raw data files in a .xlsx format.)
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
In the article we also discuss and plot data generated by Institute of Health Metrics and Evaluation26 in the 2020-06-05 update of their model. The terms and conditions can be found or their website and states for non-commercial users: “Data made available for download on IHME Websites can be used, shared, modified or built upon by non-commercial users via the Creative Commons Attribution-NonCommercial 4.0 International License (https://creativecommons.org/licenses/by-nc/4.0/)”.
The Appendix to this article as well as the R-code to generate the graphs and the nowcasts are available as extended data.
Zenodo: On the use of real-time mortality data in modelling and analysis during an epidemic outbreak – extended data.
http://doi.org/10.5281/zenodo.400524413.
This project contains the following extended data:
Liljenberg2020_OGR_Appendix.pdf (Appendix to the main article)
swedish_covid_deaths_OGR.R. (R-script to generate graphs and nowcasts in the paper.)
MDAR author checklist.pdf (Completed MDAR reporting checklist)
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We would like to acknowledge Dr. Sarah McGough for assistance with the Harvard nowcasting r-package.
| Views | Downloads | |
|---|---|---|
| Gates Open Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: infectious disease epidemiology and modeling, statistics, COVID-19, vaccination
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Sarkar K, Khajanchi S, Nieto JJ: Modeling and forecasting the COVID-19 pandemic in India.Chaos Solitons Fractals. 2020; 139: 110049 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Infectious diseases, Ecological systems, Tumor-immune interactions.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 11 Sep 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with Gates Open Research
Already registered? Sign in
If you are a previous or current Gates grant holder, sign up for information about developments, publishing and publications from Gates Open Research.
We'll keep you updated on any major new updates to Gates Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)