Keywords
Somalia, SARS-CoV-2, seroepidemiological studies, serosurvey, multi-bead assay, poliovirus, measles, rubella
This paper describes the design and methods of a serosurvey conducted in Somalia in 2021. The study had several concurrent aims: a) to estimate seroprevalence of antibodies to SARS-CoV-2, b) to obtain age-specific data on susceptibility to poliovirus, measles, rubella, diphtheria, and tetanus; and c) to estimate seroprevalence of pathogens causing malaria and neglected tropical diseases. Participants were recruited from persons seeking care at government health facilities as well as friends and family members who accompanied those seeking care. Respondents answered interview questions to establish their demographic profile, their COVID-19 exposure and experience, and, for children, their routine immunization status. Each participant contributed a sample of blood for serum or dried blood spots. Serum samples were analyzed in Somalia for SARS-CoV-2 and dried blood spots were analyzed at the US Centers for Disease Control and Prevention (US CDC) for the other diseases and antigens of interest. This manuscript describes the study design, logistics, laboratory methods, and data management steps used to compile the study dataset. Study results will be reported in a series of manuscripts to follow.
Somalia, SARS-CoV-2, seroepidemiological studies, serosurvey, multi-bead assay, poliovirus, measles, rubella
Vaccination is a cost-effective public health intervention for the control of infectious diseases. Vaccines have led to remarkable health gains over the last century and are commonly regarded as one of the greatest public health achievements in human history. Polio is currently the only vaccine-preventable disease (VPD) targeted for worldwide eradication; additionally measles is targeted for elimination in all six World Health Organization (WHO) regions. Ensuring high vaccination coverage and surveillance will be critical to polio eradication and measles elimination efforts.
Somalia is a country emerging from three decades of conflict that damaged or destroyed much of the country’s public health infrastructure1. Today, continued fragility and chronic institutional underinvestment remain significant barriers to rebuilding and strengthening the Somali healthcare system1. Nearly 70% of Somalis live in poverty today, but this is particularly severe in rural areas and among Internally Displaced Persons (IDPs) living in settlements2. Approximately 40% of Somali households live more than 30 minutes walking distance from their nearest health clinic2. As such, available sources of vaccination coverage data for Somalia are sparse and inconsistent.
According to the 2020 Somali Health and Demographic Survey, routine coverage for all the vaccines scheduled to be administered during the first year of life, specifically one Bacillus Calmette–Guérin (BCG) vaccine, three doses of pentavalent (diphtheria, pertussis, tetanus, hepatitis B, and Haemophilus influenza b) and four doses of oral polio vaccines, and one dose of measles vaccine, was only 11%. These data were verified by reference to the child vaccination card as well as caregiver’s recall when the card was missing. Only 37% of children had received BCG, 21% had received the first dose of pentavalent vaccine, and 30% had received the first dose of polio. Only 12% of children completed the required three doses of the pentavalent vaccine and 26% received three doses of oral polio vaccine. Only 23% percent of children had been vaccinated against measles and only 27% of births were protected against neonatal tetanus3. In addition to the routine doses mentioned here, oral polio and measles vaccine doses are administered through vaccination campaigns.
Vaccination coverage in Somalia is estimated by various sources, including Demographic and Health Surveys, independent post-campaign coverage surveys, administrative reports from the National Expanded Programme on Immunization (EPI), and WHO/UNICEF coverage estimates. These sources capture vaccination status but do not estimate the actual immunity of the population, in part, because natural infection and failed vaccination (due to vaccine quality, administration, or host factors such as malnutrition and infection) are not considered.
Seroepidemiological surveys (serosurveys), also known as sero-surveillance or serological surveys provide valuable insight into the natural history and epidemiology of infection and can be used to: assess the effectiveness of vaccine campaigns, determine proximity to theoretical thresholds for disease elimination, estimate the burden of VPDs, and identify gaps in population immunity to inform interventions such as supplementary immunization activities4. By estimating population susceptibility to vaccine-preventable diseases, serosurveys can provide critical insights into ongoing immunity gaps within certain sub-populations and geographic regions, as well as EPI program efficiency, especially when triangulated with surveillance and coverage data.
Somalia continues to experience cases of vaccine-derived polio virus caused by significant immunity gaps due to poor vaccination coverage to the virus. The country also reports regular outbreaks of measles, as coverage during both routine and mass vaccination campaigns has not achieved the level required to interrupt transmission (95%). Inaccessibility (e.g., difficult terrain), poor infrastructure (e.g., lack of vaccine cold chain), and chronic insecurity make vaccine delivery challenging throughout many parts of the country, resulting in sub-optimal immunity. Regardless of root cause, serosurvey data on population susceptibility to poliovirus (types 1, 2, and 3), measles, rubella, and tetanus and an improved understanding of the potential risk factors associated with insufficient population immunity may help direct appropriate programmatic actions and can provide immunity benchmarks for the elimination of these preventable diseases.
In addition, serosurveys measure immunity from vaccination or past infection. Such information is particularly helpful for mathematical modelling that can be used to determine the potential for future epidemics and at-risk age groups4. This knowledge can help determine the need for public health intervention by estimating the true level of population immunity to antigens of interest.
In 2021, the WHO undertook a serosurvey in Somalia to estimate seroprevalence of antibodies to SARS-CoV-2 and a long list of other diseases. This paper describes the survey methods, sample processing methods, and data management steps employed to construct the analysis dataset that combines interview responses with three sets of laboratory results.
The aims of the survey were to:
1. estimate seroprevalence of antibodies to SARS-CoV-2;
2. obtain age-specific data on susceptibility to poliovirus, measles, rubella, diphtheria, and tetanus;
3. identify potential risk factors associated with insufficient population immunity to vaccine preventable diseases;
4. identify areas at high risk for polio outbreaks;
5. identify areas where immunization coverage remains low;
6. estimate seroprevalence of pathogens causing malaria and neglected tropical diseases.
The study was a cross-sectional population-based survey conducted with a serosurvey that was part of the WHO Unity COVID-19 studies5–7. Each participant completed a questionnaire and had a sample of venous blood collected. Data were collected from March to July 2021.
The study was conducted across Somalia, including the Banadir Regional Administration, Puntland, and Somaliland, but excluding Middle Juba for security and operational reasons. Eligible respondents were persons ages 1 year and older who were visiting non-respiratory disease outpatient or inpatient departments of public health facilities (to reduce potential confounding for the COVID-19 section of this study). Accompanying household members were also eligible; however, only one person per household was enrolled.
Exclusion criteria included refusal or inability to provide informed consent, contraindication to or unsuccessful venipuncture after three attempts, and individuals with critical illnesses or undergoing serious medical intervention.
The study was conducted in accordance with the World Medical Association’s Declaration of Helsinki on ethical principles for medical research involving human subjects8. There was no formal ethical review board in Somalia at the time the work was initiated but the study protocol was approved by the Somali Health Authority on 11 November 2020. The health facility authorities and outpatient team members at data collection sites were informed of the study and its objectives in advance to facilitate safe and efficient data collection without interfering with facility operations.
This activity was reviewed by CDC, deemed not research, and was conducted consistent with applicable federal law and CDC policy (See e.g., 45 C.F.R. part 46, 21 C.F.R. part 56; 42 U.S.C. §241(d); 5 U.S.C. §552a; 44 U.S.C. §3501 et seq.).
The study target sample size was 3,616 respondents. The inferential goal was to yield state-level two-sided Wald type 97% confidence intervals no wider than +/- 5% for estimated proportions of 50%, on average, if the data were analyzed as a simple random sample with a nonresponse or missing results rate of 5% of respondents. The sample target was allocated across administrative regions in proportion to estimated population (Table 1).
In total, 19 administrative regions were visited to collect data at the regional hospital and at every district health center. Each data collection team was asked to achieve a balance of male and female respondents and to allocate the sample across five age groups in quotas designed to mirror the age distribution of the Somali population. Each team was assigned a target number of respondents for each of five age groups. When aggregated, the sample was designed to be distributed as follows: 20.3% ages 1–4; 34.5% ages 5 to 14; 22.9% ages 15 to 29; 13.1% ages 30 to 49; and 9.2% age 50 and above.
Family groups utilizing the health facility services were approached for enrollment. A table of random numbers was used to identify the first group to approach and thereafter every fifth or tenth family was approached (depending on the volume of persons seeking care) until the target number of respondents was recruited. Within a selected family, a single respondent was chosen using simple random sampling. If that person’s age group quota had already been filled, another family member was selected from an age group that had not been filled.
Paper informed consent forms were signed by eligible respondents before the study team administered the questionnaire and collected specimens. Parental consent was obtained for children under 17 years, and assent was obtained from children between 7 and 17 years. Participants were informed that they would be tested for the presence of VPD antibodies and COVID-19 antibodies. Every participant could ask questions throughout the process. The study procedures were described along with risks and benefits, and it was clearly explained that their decision whether to join the study would not affect how they were treated at the clinic.
Small gifts (worth no more than $2) were given to those who participated. Children who were found to have missed some vaccinations were referred for an opportunity to receive them.
The survey was implemented using ODK9,10 and data were recorded using the interviewers’ personal smartphones. Figure 1 and Figure 2 are maps that show the sites where data were collected. Figure 3 shows the timeline of data collection and lab result availability. The questionnaire consisted of several short modules, some of which were limited to a subset of participants, as shown in Table 2. The ODK questionnaire is available in XLSForm format from an online repository11,12. Every participant was categorized as either an adult (age 18 and above and answering on behalf of themselves) or a guardian (answering on behalf of a child ages 1–17 years). Barcode stickers were affixed to the paper consent forms and then scanned with ODK to serve as a unique participant identifier.
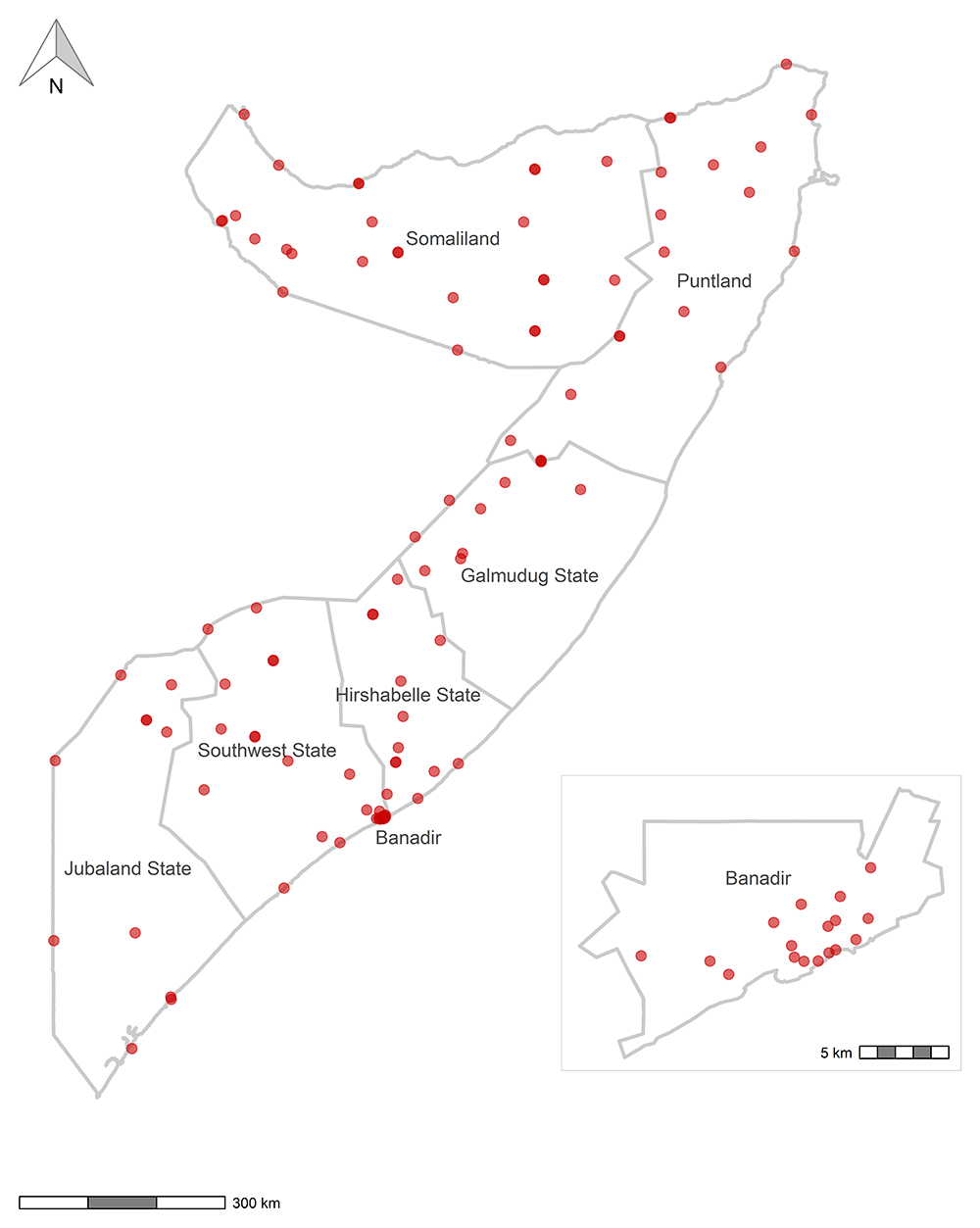
Note: No data were collected in Middle Juba in Jubaland State because of insecurity and unrest. Note: The boundaries and names shown and the designations used on these maps do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted and dashed lines on maps represent approximate border lines for which there may not yet be full agreement. Data source: WHO Somalia 2021 Serosurvey. Map production: Biostat Global Consulting. World Health Organization © WHO 2023. All rights reserved.
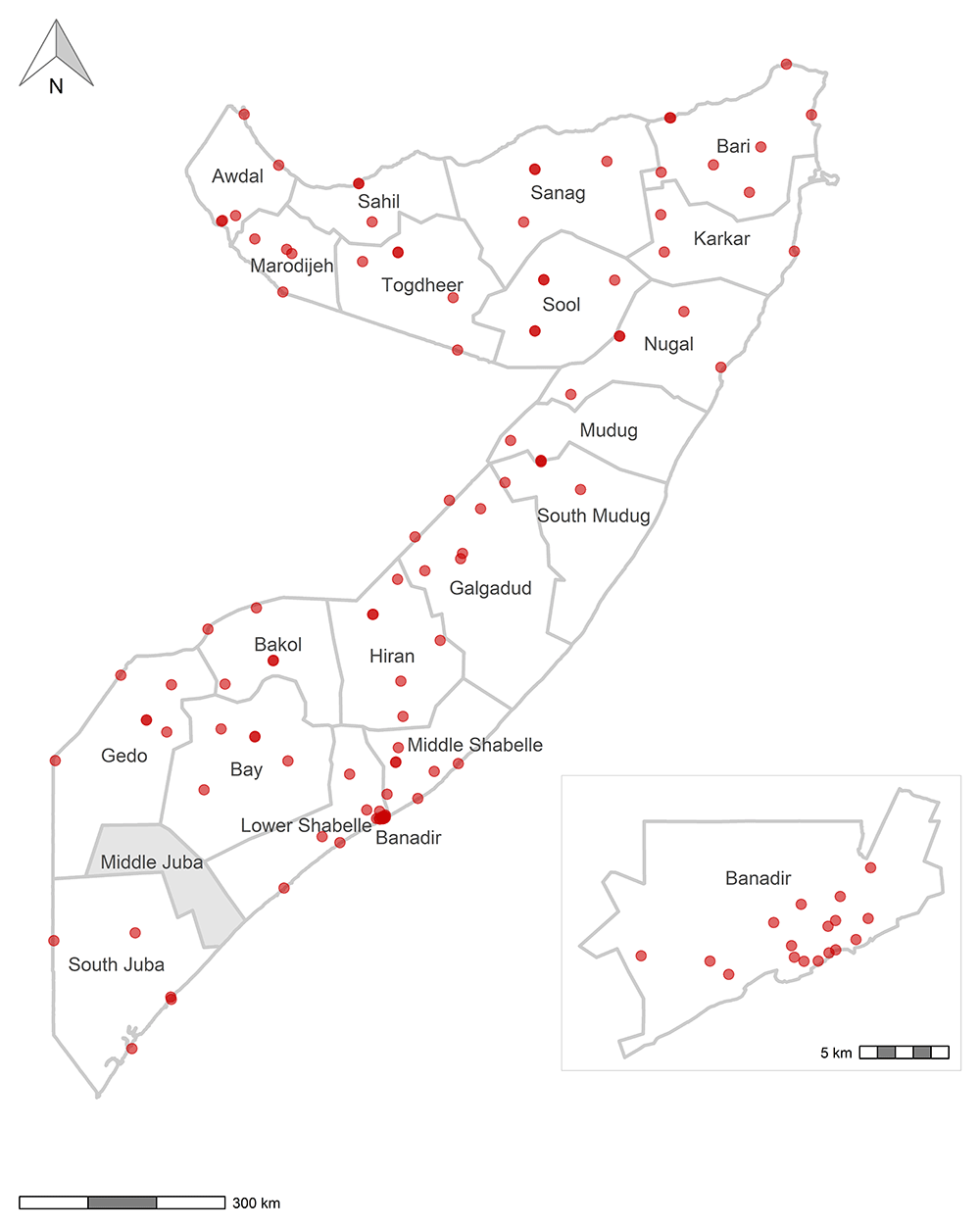
Note: No data were collected in Middle Juba in Jubaland State because of insecurity and unrest. Note: The boundaries and names shown and the designations used on these maps do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted and dashed lines on maps represent approximate border lines for which there may not yet be full agreement. Data source: WHO Somalia 2021 Serosurvey. Map production: Biostat Global Consulting. World Health Organization © WHO 2023. All rights reserved.
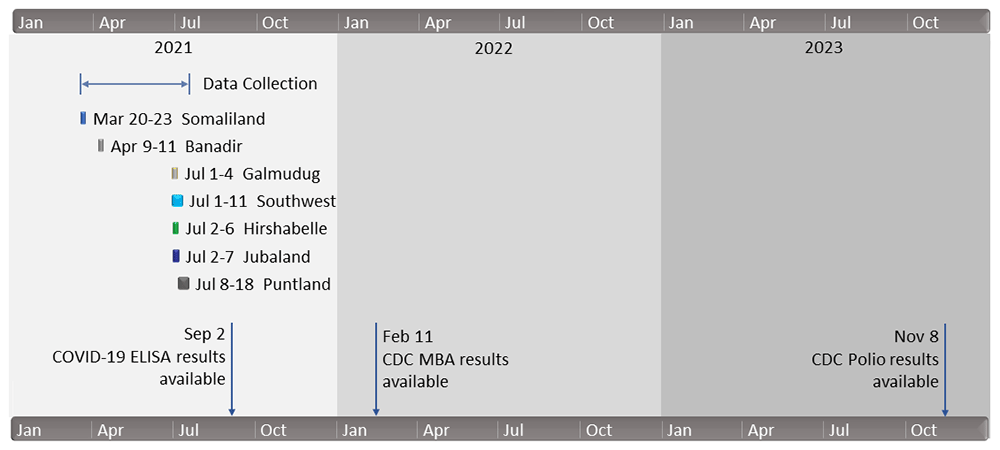
Note: Poliovirus assay results were delayed by a) a global shortage of supplies for the assay, b) scheduling conflicts in relation to other studies at the US CDC Polio and Picornavirus Laboratory Branch, and c) problems with individual DBS cards described in the text.
The survey was conducted in Somaliland first, and some ambiguities concerning the ages of adult and guardian participants were quickly identified in the dataset. The survey form was updated to include additional demographic questions and to solve the problem of ambiguous ages. The form completed in subsequent interviews therefore had minor changes from the Somaliland form. Respondents in the subsequent interviews were asked some questions that were not posed in Somaliland, so those variables have been coded “Not Asked” for Somaliland participants.
Trained phlebotomists collected 5 ml venous blood samples in red-top (no additives) test-tubes and handled them according to the WHO Unity study protocol. Dried blood spot (DBS) samples were prepared using prepacked DBS kits. From each participant, five blood spots between 300 and 500µl were placed on filter paper using a dropper. DBS samples were left to dry on a rack for at least three hours before being packaged with desiccant and a humidity indicator card before storage at the Somalia national laboratory in Mogadishu and then combined with all samples from data collection locations and shipped to the US CDC in Atlanta for analysis. Barcode stickers that matched the one applied to the consent form were affixed to each respondent’s test tube and DBS card. When possible, regional and district-level supervisors from WHO were present to monitor the quality of the work.
The processes for serum sample handling and analysis are described elsewhere13. Labeled tubes were kept at 2–8°C and transferred to regional hospital laboratories where they were separated and transferred again, still at 2–8°C to three regional reference laboratories (in Mogadishu, Garowe, and Hargeisa) where they were analyzed using the WANTAI SARS COV-2 Ab enzyme-linked immunosorbent assay (ELISA) according to manufacturer's instructions (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd, China). Transfer to the reference laboratory was achieved within one day of collection for most samples. Due to uncertain travel conditions resulting from political strife, some samples from South & Central Somalia were stored as long as five days, still at 2–8°C, before arriving at the destination laboratory. The kit has a combined IgM and IgG specificity of 97.5% and sensitivity of 96.7% for symptomatic COVID-19 cases14,15. The ELISA results (positive, negative or non-conclusive) were entered into the study data repository using ODK in a format amenable to merging with survey responses using the barcode as a unique matching key.
DBS cards were analyzed first using MBA1 to evaluate seropositivity for 27 antigens associated with 12 diseases using a small punch from a dried blood spot. See Table 3 and Table 4. The MBA is a multiplex assay that allows antibodies for multiple pathogens to be tested simultaneously for a sample in a single well using small sample volumes16. Although the gold standard for evaluating seroprotection against measles is the plaque reduction neutralization (PRN) assay, and the focus reduction assay for rubella, the MBA has been validated as an acceptable alternative16,17. MBAs have been used in other recent serosurveys for measles, mumps, rubella, tetanus, diphtheria, malaria, and yaws18–25.
| Pathogen / Source Organism | Antigen | Abbrev. | GenBank accession number or Strain | References | Multiplex References | Coupling Conc (µg / 1.25 e7 beads | Coupling pH | Antigen type and source |
|---|---|---|---|---|---|---|---|---|
| Schistosoma japonicum | Glutathione-S-transferase | GST | 27,28 | 29 | 15 | 5 | Recombinant, CDC/ LSDB | |
| Cercopithecus aethiops | Vero cell lysate | Vero | CCL-81 | 16 | 150 | 7.2 | Native, CDC / VVPDB | |
| Plasmodium falciparum 3D7 | Merozoite surface protein 1, 19-kDa subunit | PfMSP119 | M64681 | 30–32 | 33,34 | 20 | 5 | Recombinant, CDC/ LSDB |
| Plasmodium vivax Belem | Merozoite surface protein 1, 19-kDa subunit | PvMSP119 | AF435594.1 | 35,36 | 37 | 20 | 5 | Recombinant, CDC/ LSDB |
| Plasmodium falciparum | Circumsporozoite protein repeat peptide (NANP)5 | PfCSP | M83173 | 38,39 | 37 | 30 | 5 | Synthesized peptide, CDC/ LSDB |
| Plasmodium falciparum FVO | Apical membrane antigen 1, ectodomain | PfAMA1 | U84348 | 40 | 41 | 20 | 5 | Recombinant, CDC/ LSDB |
| Plasmodium falciparum Type A and B (1:1) | Histidine rich protein 2 | PfHRP2 | CZT62760.1 | 42,43 | 41,44 | 25 | 5 | Recombinant, ICL Labs, Portland, OR |
| Plasmodium falciparum | Liver surface antigen 1 | PfLSA1 | Pl1043 epimiddlee | 45 | 41,46 | 60 | 5 | Synthesized peptide, CDC/ LSDB |
| Plasmodium falciparum | Glutamate rich protein R0 | PfGLURP R0 | R0 fragment | 57 | 41 | 30 | 5 | Synthesized peptide, CDC/ LSDB |
| Strongyloides stercoralis | NIE | AAB97359 | 48 | 49 | 20 | 7.2 + Urea | Recombinant, CDC/ LSDB | |
| Chlamydia trachomatis | pCT03 ORF | Pgp3 | 50 | 51 | 120 | 7.2 | Recombinant, CDC/ LSDB | |
| Brugia malayi | SXP-1 | Bm14 | M95546 | 52 | 53 | 120 | 7.2 | Recombinant, CDC/ LSDB |
| Brugia malayi | Pepsin inhibitor analog AP-1 | Bm33 | L11001 | 54 | 55 | 20 | 6 + Urea | Recombinant, CDC/ LSDB |
| Wucheraria bancrofti | Larval antigen | Wb123 | HQ438580 | 56,57 | 26 | 30 | 7.2 | Recombinant, NIH/ T. Nutman |
| Onchocerca volvulus | Ag16 Diagnostic antigen | Ov16 | M27807 | 58,59 | 60 | 30 | 7.2 | Recombinant, NIH/ T. Nutman |
| Onchocerca volvulus | Pepsin inhibitor analog | Ov33 | X13313.1 | 61,62 | 60 | 30 | 5 | Recombinant, CDC/ LSDB |
| Schistosoma mansoni | Soluble egg antigen | SEA | S. mansoni | 63 | 64 | 120 | 7.2 | Native, CDC/ LSDB |
| Schistosoma mansoni | Sm25 | M37004.1 | 65,66 | 64 | 12 | 7.2 | Recombinant, CDC/ LSDB | |
| Clostridium tetani | Tetanus toxoid | TetTox | Com. product | 67,68 | 69,70 | 12.5 | 5 | Native, Massachusetts Biologic Laboratories, Boston, MA |
| Corynebacterium diphtheriae | Diphtheria toxoid | DipTox | Com. product | 68,71,72 | 73,74,75 | 60 | 5 | Native, List Biological Labs, Campbell, CA |
| Rubella virus | Whole virus | wRuV | Com. product | 76 | 16,77,78 | 30 | 5 | Native, Meridian Life Sciences, Memphis, TN |
| Measles virus | Whole virus | wMeV | Com. product | 80 | 16 | 150 | 7.2 | Native, ZeptoMetrix, Buffalo, NY |
| Treponema pallidum | Treponemal membrane protein A (Yaws) | TmpA | Com. product | 79,80 | 24 | 30 | 7.2 | Recombinant, Virogen, Watertown, MA |
| Treponema pallidum | Recombinant protein 17 | rp17 | Com. product | 81 | 24 | 30 | 7.2 | Recombinant, Virogen, Watertown, MA |
| SARS-CoV-2 | Spike full length trimer | Spike | MN908947 | 82 | 83 | 6 | 7.2 | Recombinant, CDC/ CORVLB |
| SARS-CoV-2 | Spike receptor binding domain residues 319-541 | RBP541 | MN908947 | 83 | 15 | 5 | Recombinant, CDC/ CORVLB | |
| SARS-CoV-2 | Spike receptor binding domain residues 319-591 | RBP591 | MN908947 | 83 | 6 | 5 | Recombinant, CDC/ CORVLB | |
| SARS-CoV-2 | Nucelocapsid protein | N | P0DTC9 | 82 | 83 | 3 | 7.2 | Recombinant, GenScript, Piscataway, NJ |
5 = 50 mM MES, 0.85% NaCl at pH 5; 7.2 = 1x phosphate buffered saline pH 7.2; 6 + 2 M Urea = 25 mM MES, 200 mM NaCl, 2 M Urea at pH 6; 7.2 + Urea = 50 mM NaH2PO4, 200 mM NaCl, 2M Urea at pH 7.2
VVPDB = Laboratory Science and Diagnostics Branch; CORVLB = Coronavirus and other Respiratory Viruses Laboratory Branch; LSDB = Laboratory Science and Diagnostics Branch; NIH = National Institutes of Health
Antigens were coupled to MagPlex microspheres (Luminex Corp., Austin, TX) as previously described26; each antigen had been previously optimized to the appropriate coupling concentration and pH summarized in Table 4. In addition to antigens, the glutathione-S-transferase (GST) and Vero cell lysate were included as non-immunogenic proteins to correct for potential non-specific binding to recombinant proteins and measles and rubella antigens, respectively.
DBS were diluted to a final estimated serum concentration of 1:400 by first eluting a 6 mm DBS punch overnight at 4°C in 200 µL of Buffer B (1X PBS, 0.5% casein, 0.5% polyvinyl alcohol [PVA], 0.8% polyvinylpyrrolidone [PVP], 0.3% Tween-20, 0.02% sodium azide and 3 μg/mL Escherichia coli extract) followed by a second dilution of 20 µl eluate into 180 µl of Buffer B.
For the MBA, all incubation steps were performed at room temperature in 50 µl reaction volumes protected from light while shaking at 600 rpm. Each incubation was followed by three washes of 200 µl PBS pH 7.2 containing 0.05% Tween 20 (PBST) using a 405-TSRS automated plate washer with a magnetic adapter (BioTek, Winooski, VT, USA). Diluted sera were incubated with ~1000 microspheres/antigen/well for 90 minutes, followed by a 45-minute incubation with secondary antibodies diluted to 50 ng/well for anti-human IgG and 40 ng/well for anti-IgG4 (Southern Biotech, Birmingham, AL) in Buffer A (1X PBS, 0.5% BSA, 0.05% Tween-20, and 0.02% NaN3). Next, plates were incubated for 30 minutes with 250 ng/well streptavidin-R phycoerythrin (Invitrogen, Waltham, MA, USA) diluted in Buffer A followed by an incubation in Buffer A alone for 30 minutes to remove loosely bound antibody complexes. Finally, microspheres were resuspended in 100 µl PBS pH 7.2 and stored overnight at 4°C prior to reading on MAGPIX (Luminex Corporation, Austin, TX, USA).
Data were output as median fluorescence intensity (MFI). To control for background reactivity, each assay included blank wells containing Buffer B only. Samples were run in single wells. Each plate contained one serum control negative for most infectious diseases and three controls with positive reactions for all antigens included in the assay. As a measure of plate-to-plate variation, the average reactivities of the five antigen-control pairs was used to create criteria for accepting or rejecting plate data.
Different approaches were used to establish seropositivity thresholds for each antigen: (1) the mean of the natural log MFI distribution of non-endemic negative controls plus three standard deviations; (2) receiver operating characteristic (ROC) curve of MFI values for negative and positive controls, using Youden’s J-index of the ROC curve to balance sensitivity and specificity; (3) for measles, rubella, diphtheria, and tetanus antigens only: interpolation of MFI into international units using a five-parameter logistic curve fit to standard dilution series. Seroprotective thresholds were based on previously validated IU values16,69,73. Methods used for each antigen are summarized in Table 5.
| Pathogen | Abbrev. | Cutoff Method References | Cutoff Method | Negative Panel | Positive Panel |
|---|---|---|---|---|---|
| Plasmodium falciparum 3D7 | PfMSP119 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium vivax Belem | PvMSP119 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium falciparum | PfCSP | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium falciparum FVO | PfAMA1 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium falciparum Type A and B | PfHRP2 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium falciparum | PfLSA1 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Plasmodium falciparum | PfGLURP R0 | 33 | LN transformed mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Strongyloides stercoralis | NIE | 49 | ROC Curve | Adults from non-endemic region (n=85) | Larvae positive adults from Argentina (n=53) |
| Chlamydia trachomatis | Pgp3 | 51,86 | ROC Curve | 1-9 year olds from New York (n=74) | Trachoma NAAT positive 1-9 year olds (n=95) |
| Chlamydia trachomatis | CT694 | 51,86 | ROC Curve | 1-9 year olds from New York (n=74) | Trachoma NAAT positive 1-9 year olds (n=95) |
| Brugia malayi | Bm14 | 26 | mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Brugia malayi | Bm33 | 26 | mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Wucheraria bancrofti | Wb123 | 26 | mean + 3 stdev | Adults from non-endemic region (n=85) | n/a |
| Onchocerca volvulus | Ov16 | 60 | ROC Curve | Adults from non-endemic region (n=57) | Skin snip microfilaria positive individuals endemic African countries (n=33) |
| Onchocerca volvulus | Ov33 | 60 | ROC Curve | Adults from non-endemic region (n=57) | Skin snip microfilaria positive individuals endemic African countries (n=33) |
| Schistosoma mansoni | SEA | ROC Curve | Adults from non-endemic region (n=131) | S. mansoni (n=72), S. haematobium (n=45) and S. intercalatum (n=77) positive individuals | |
| Schistosoma mansoni | Sm25 | ROC Curve | Adults from non-endemic region (n=131) | S. mansoni (n=72) positive individuals | |
| Clostridium tetani | TetTox | 69,70 | WHO Standard Curve TE-3 | n/a | n/a |
| Corynebacterium diphtheriae | DipTox | 73,74,75 | WHO Standard Curve 10/262 | n/a | n/a |
| Rubella virus | wRuV | 16,77,78 | WHO Standard Curve 67/182 | n/a | n/a |
| Measles virus | wMeV | 16,77 | WHO Standard Curve 97/648 | n/a | n/a |
| Treponema pallidum | TmpA | 24 | ROC Curve | 1-9 year-olds from New York (n=74), RPR negative 4-15 year-olds from Vanuatu (n=65) | TPPA positive 4-15 year-olds from Vanuatu (n=36) |
| Treponema pallidum | rp17 | 24 | ROC Curve | 1-9 year-olds from New York (n=74); TPPA negative 4-15 year-olds from Vanuatu (n=42) | TPPA positive 4-15 year-olds from Vanuatu (n=36) |
| SARS-CoV-2 | Spike | 83 | ROC Curve | Plasma collected prior to Nov 2019 (n=99); RT-PCR negative, Mar-June 2020 (n=19) | Plasma collected in Mar-June 2020, RT-PCR positive (n=87) |
| SARS-CoV-2 | RBP541 | 83 | ROC Curve | Plasma collected prior to Nov 2019 (n=99); RT-PCR negative, Mar-June 2020 (n=19) | Plasma collected in Mar-June 2020, RT-PCR positive (n=87) |
| SARS-CoV-2 | RBP591 | 83 | ROC Curve | Plasma collected prior to Nov 2019 (n=99); RT-PCR negative, Mar-June 2020 (n=19) | Plasma collected in Mar-June 2020, RT-PCR positive (n=87) |
| SARS-CoV-2 | N | 83 | ROC Curve | 99 plasma collected prior to Nov 2019; 19 RT-PCR negative, Mar-June 2020 | 87 plasma collected in Mar-June 2020, RT-PCR positive |
A poliovirus microneutralization assay was used to assess levels of neutralizing antibodies against all three poliovirus serotypes84. For this study, levels of poliovirus neutralizing antibodies against poliovirus serotypes 1, 2, and 3 were assessed from DBS84 collected on a Whatman Protein Saver Card85. Samples were randomized using a balanced block randomization scheme with integrated controls using MATLAB84. Each serum was tested in triplicate using step 1:2 dilutions ranging from 1:8-1:1024. The cutoff for neutralization was determined by calculating 80% of the average luminescence from cell control wells within each test plate. Each test run included back titration and serum (in-house reference human serum with known levels of poliovirus neutralizing antibodies). Seropositivity was defined as the reciprocal titer of poliovirus-neutralizing antibodies ≥8.
Figure 4 shows how the data sources moved through data collection, laboratory testing, and then into the analysis dataset. Note that for a variety of reasons, some respondents did not have some lab results to merge with their interview responses.
Each interview participant was assigned four survey weights. A population weight was assigned by region and age group, so the sum of weights would equal the population figures used to plan the study. Everyone in the analysis dataset had a positive population weight. MBA weights were assigned to maintain the sums of weights by state and by age group for persons whose interview data could be merged with their MBA results. Persons with no MBA results had an MBA weight of 0, and their population weights were spread across or shifted to the persons in their same state and age group who did have MBA results. Everyone with MBA results had a positive MBA weight. Similarly, everyone with COVID-19 ELISA results had a positive serum weight and everyone with polio assay results had a positive polio weight. For MBA and serum and polio weights, the weights of persons without merged results were spread equally across all the respondents in their same state and age group who did have results. Note that none of the weights attempt to correct imbalance for sex of the respondents or for the mix of urban and rural respondents; the only constructs that were incorporated into population weights were region and age group, and for the adjusted MBA, serum, and polio weights, state, and age group.
When the study was planned, the Bari region was a combination of what might now be called the Bari and Karkar regions. The populations for what are now two regions were combined. Data were collected at health facilities in both regions. If analysts wish to calculate separate results for Karkar, then their code should change the value of the region code for participants from these three facilities from Bari to Karkar: Qardho District Hospital, Waciye Health Center, and Bayla Referral Health Center.
The sample demographics, seroprevalence results and other work products will be described in publications to follow.
The methodology used in this study has several strengths. It used the WHO Unity Studies protocol and therefore is meaningfully comparable to other such studies. It includes respondents from all around Somalia (except for the Middle Juba region) and from a wide variety of ages. Missingness among lab results was partially corrected by reweighting the respondents whose results were present. This study also highlights the use of DBS as an effective technique especially in a resource-poor setting and when surveying hard-to-reach populations such as those in Somalia. In settings where collection, storage, transport, and timely processing of serum can be challenging, DBS can be an especially viable and effective alternative that can be used for many different infections.
The study methodology also has limitations. Persons who did not seek care or accompany those who sought care at government health facilities would not have been recruited into the sample. Insecurity at and between some study sites prevented having a full complement of supervisors present on every study day, so some quality checks were missed. The list of survey questions was updated to include more internal consistency reviews after data were collected in Somaliland and Banadir. Some of the barcodes that were supposed to facilitate unambiguous merging of interview data with lab results were mismanaged, yielding some samples that had identical bar codes and some samples whose codes did not match any of those from the interview responses. This led to missing lab results. If the missingness was due to varying levels of care and competence across the data collection teams, then the missingness was not completely at random. Further, despite careful processing, some samples were not analyzable, having been affected by mold or other contaminants. Hence the results might be interpreted as indicative, if not representative, of the study sample or the Somali population.
Zenodo: Somalia Serosurvey 2021 – VPD and COVID-19 Questionnaire, https://doi.org/10.5281/zenodo.1064054812.
First, the authors thank all the study participants for engaging with the data collection teams and contributing to scientific knowledge in their communities. Further, this study would not have been possible without instrumental contributions of the Minister of Health and Human Services, Federal Government of Somalia, the Federal and State Ministry of Health and WHO COVID-19 Incident Management support team members, the District and Regional Medical Officers of the Ministry of Health, the District, Regional and State Public Health Emergency Officers of the WHO in Somalia, WHO EPI team members, the enumerators, phlebotomists and laboratory technicians collecting the data and samples in the health facilities, laboratory technicians working in Mogadishu National Public Health Reference Laboratory, Puntland State Public Health Laboratory and Somaliland Public Health Laboratory and the USA CDC MBA and Polio (Basit Jafri, Kathryn Jones, Eric Rhoden, Giovanna Sifontes, Sandra Valdez, Nicholas Wiese, and Yiting Zhang and the Specimen Triage and Tracking Team (STATT)) assay teams.
The findings in this article are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
| Views | Downloads | |
|---|---|---|
| Gates Open Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Partly
Are sufficient details provided to allow replication of the method development and its use by others?
Partly
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: I am employed by the Gates Foundation, but in a different part of the foundation from the study body that would have awarded the grant.
Reviewer Expertise: Infectious disease epidemiology. I am not as familiar with laboratory methods but it seems the first reviewer has already looked over those in detail.
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Partly
Are sufficient details provided to allow replication of the method development and its use by others?
Partly
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Epidemiology, Disease Control, Serosurveillance
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Partly
Are sufficient details provided to allow replication of the method development and its use by others?
Partly
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: humoral and mucosal immunology, serosurveillence
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 01 Mar 24 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Register with Gates Open Research
Already registered? Sign in
If you are a previous or current Gates grant holder, sign up for information about developments, publishing and publications from Gates Open Research.
We'll keep you updated on any major new updates to Gates Open Research
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)